Lesson type
Difficulty level
This lesson provides an overview of the current status in the field of neuroscientific ontologies, presenting examples of data organization and standards, particularly from neuroimaging and electrophysiology.
Difficulty level: Intermediate
Duration: 33:41
Speaker: : Yaroslav O. Halchenko
The lesson introduces the Brain Imaging Data Structure (BIDS), the community standard for organizing, curating, and sharing neuroimaging and associated data. The session focuses on understanding the BIDS framework, learning its data structure and validation processes.
Difficulty level: Intermediate
Duration: 38:52
Speaker: : Cyril Pernet
This session moves from BIDS basics into analysis workflows, focusing on how to turn raw, BIDS-organized data into derivatives using BIDS Apps and containers for reproducible processing. It compares end-to-end pipelines across fMRI and PET (and notes EEG/MEG), explains typical preprocessing choices, and shows how standardized inputs plus containerized tools (Docker/AppTainer) yield consistent, auditable outputs.
Difficulty level: Intermediate
Duration: 56:03
Speaker: : Martin Nørgaard
The session explains GDPR rules around data sharing for research in Europe, the distinction between law and ethics, and introduces practical solutions for securely sharing sensitive datasets. Researchers have more flexibility than commonly assumed: scientific research is considered a public interest task, so explicit consent for data sharing isn’t legally required, though transparency and informing participants remain ethically important. The talk also introduces publicneuro.eu, a controlled-access platform that enables sharing neuroimaging datasets with open metadata, DOIs, and customizable access restrictions while ensuring GDPR compliance.
Difficulty level: Intermediate
Duration: 31:12
Speaker: : Cyril Pernet
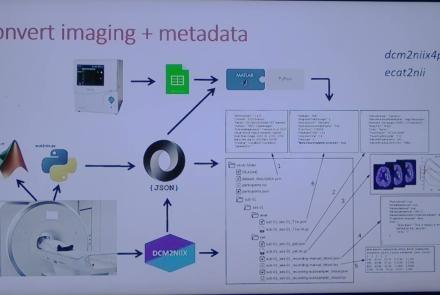
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 41:04
Speaker: : Martin Nørgaard
This session dives into practical PET tooling on BIDS data—showing how to run motion correction, register PET↔MRI, extract time–activity curves, and generate standardized PET-BIDS derivatives with clear QC reports. It introduces modular BIDS Apps (head-motion correction, TAC extraction), a full pipeline (PETPrep), and a PET/MRI defacer, with guidance on parameters, outputs, provenance, and why Dockerized containers are the reliable way to run them at scale.
Difficulty level: Intermediate
Duration: 1:05:38
Speaker: : Martin Nørgaard
This session introduces two PET quantification tools—bloodstream for processing arterial blood data and kinfitr for kinetic modeling and quantification—built to work with BIDS/BIDS-derivatives and containers. Bloodstream fuses autosampler and manual measurements (whole blood, plasma, parent fraction) using interpolation or fitted models (incl. hierarchical GAMs) to produce a clean arterial input function (AIF) and whole-blood curves with rich QC reports ready. TAC data (e.g., from PETPrep) and blood (e.g., from bloodstream) can be ingested using kinfitr to run reproducible, GUI-driven analyses: define combined ROIs, calculate weighting factors, estimate blood–tissue delay, choose and chain models (e.g., 2TCM → 1TCM with parameter inheritance), and export parameters/diagnostics. Both are available as Docker apps; workflows emphasize configuration files, reports, and standard outputs to support transparency and reuse.
Difficulty level: Intermediate
Duration: 1:20:56
Speaker: : Granville Matheson
This lecture covers the NIDM data format within BIDS to make your datasets more searchable, and how to optimize your dataset searches.
Difficulty level: Beginner
Duration: 12:33
Speaker: : David Keator
This lecture covers positron emission tomography (PET) imaging and the Brain Imaging Data Structure (BIDS), and how they work together within the PET-BIDS standard to make neuroscience more open and FAIR.
Difficulty level: Beginner
Duration: 12:06
Speaker: : Melanie Ganz
This lecture discusses how to standardize electrophysiology data organization to move towards being more FAIR.
Difficulty level: Beginner
Duration: 15:51
Speaker: : Sylvain Takerkart
Hierarchical Event Descriptors (HED) fill a major gap in the neuroinformatics standards toolkit, namely the specification of the nature(s) of events and time-limited conditions recorded as having occurred during time series recordings (EEG, MEG, iEEG, fMRI, etc.). Here, the HED Working Group presents an online INCF workshop on the need for, structure of, tools for, and use of HED annotation to prepare neuroimaging time series data for storing, sharing, and advanced analysis.
Difficulty level: Beginner
Duration: 03:37:42
Speaker: :
In this lesson, attendees will learn about the data structure standards, specifically the Brain Imaging Data Structure (BIDS), an INCF-endorsed standard for organizing, annotating, and describing data collected during neuroimaging experiments.
Difficulty level: Beginner
Duration: 21:56
Speaker: : Michael Schirner
This is the first of two workshops on reproducibility in science, during which participants are introduced to concepts of FAIR and open science. After discussing the definition of and need for FAIR science, participants are walked through tutorials on installing and using Github and Docker, the powerful, open-source tools for versioning and publishing code and software, respectively.
Difficulty level: Intermediate
Duration: 1:20:58
Speaker: : Erin Dickie and Sejal Patel
Course:
In this lesson, while learning about the need for increased large-scale collaborative science that is transparent in nature, users also are given a tutorial on using Synapse for facilitating reusable and reproducible research.
Difficulty level: Beginner
Duration: 1:15:12
Speaker: : Abhi Pratap
This lesson contains the first part of the lecture Data Science and Reproducibility. You will learn about the development of data science and what the term currently encompasses, as well as how neuroscience and data science intersect.
Difficulty level: Beginner
Duration: 32:18
Speaker: : Ariel Rokem
In this second part of the lecture Data Science and Reproducibility, you will learn how to apply the awareness of the intersection between neuroscience and data science (discussed in part one) to an understanding of the current reproducibility crisis in biomedical science and neuroscience.
Difficulty level: Beginner
Duration: 31:31
Speaker: : Ashley Juavinett
Course:
The lecture provides an overview of the core skills and practical solutions required to practice reproducible research.
Difficulty level: Beginner
Duration: 1:25:17
Speaker: : Fernando Perez
Course:
This lecture provides an introduction to reproducibility issues within the fields of neuroimaging and fMRI, as well as an overview of tools and resources being developed to alleviate the problem.
Difficulty level: Beginner
Duration: 1:03:07
Speaker: : Russell Poldrack
Course:
This lecture provides a historical perspective on reproducibility in science, as well as the current limitations of neuroimaging studies to date. This lecture also lays out a case for the use of meta-analyses, outlining available resources to conduct such analyses.
Difficulty level: Beginner
Duration: 55:39
Speaker: : Angela Laird
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)