Course:
In this lesson, while learning about the need for increased large-scale collaborative science that is transparent in nature, users also are given a tutorial on using Synapse for facilitating reusable and reproducible research.
Difficulty level: Beginner
Duration: 1:15:12
Speaker: : Abhi Pratap
This lecture discusses what defines an integrative approach regarding research and methods, including various study designs and models which are appropriate choices when attempting to bridge data domains; a necessity when whole-person modelling.
Difficulty level: Beginner
Duration: 1:28:14
Speaker: : Dan Felsky
This lesson provides an introduction the International Neuroinformatics Coordinating Facility (INCF), its mission towards FAIR neuroscience, and future directions.
Difficulty level: Beginner
Duration: 20:29
Speaker: : Maryann Martone
This brief video provides an introduction to the third session of INCF's Neuroinformatics Assembly 2023, focusing on how to streamling cross-platform data integration in a neuroscientific context.
Difficulty level: Beginner
Duration: 5:55
Speaker: : Bing-Xing Huo
This final lesson of the course consists of the panel discussion for Streamlining Cross-Platform Data Integration session during the first day of INCF's Neuroinformatics Assembly 2023.
Difficulty level: Beginner
Duration: 50:16
Speaker: :
This lightning talk describes the heterogeneity of the MR field regarding types of scanners, data formats, protocols, and software/hardware versions, as well as the challenges and opportunities for unifying these datasets in a common interface, MRdataset.
Difficulty level: Beginner
Duration: 5:15
Speaker: : Harsh Sinha
This session covers the framework of the International Brain Lab (IBL) and the data architecture used for this project.
Difficulty level: Beginner
Duration: 23:37
Speaker: : Kenneth Harris
This lecture covers a lot of post-war developments in the science of the mind, focusing first on the cognitive revolution, and concluding with living machines.
Difficulty level: Beginner
Duration: 2:24:35
Speaker: : Paul F.M.J. Verschure
This brief talk goes into work being done at The Alan Turing Institute to solve real-world challenges and democratize computer vision methods to support interdisciplinary and international researchers.
Difficulty level: Beginner
Duration: 7:10
Speaker: : Alden Connor & Beatriz Costa Gomes
This lesson aims to define computational neuroscience in general terms, while providing specific examples of highly successful computational neuroscience projects.
Difficulty level: Beginner
Duration: 59:21
Speaker: : Alla Borisyuk
Course:
This lecture gives an introduction to simulation, models, and the neural simulation tool NEST.
Difficulty level: Beginner
Duration: 1:48:18
Speaker: : Marc-Oliver Gewaltig
Course:
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lesson discuses forms of neural plasticity on many levels, including short-term, long-term, metaplasticity, and structural plasticity. During the lesson you will also be presented with examples related to the modelling of biochemical networks.
Difficulty level: Beginner
Duration: 1:11:29
Speaker: : Upi Bhalla
This lesson provides an introduction to modelling of chemical computation in the brain.
Difficulty level: Beginner
Duration: 1:00:11
Speaker: : Upi Bhalla
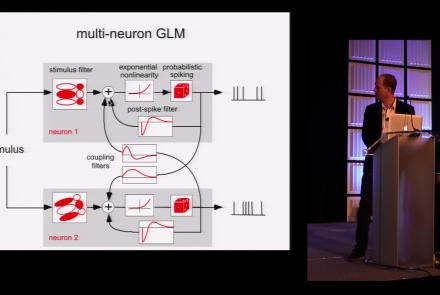
This lesson is part 1 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:45:48
Speaker: : Jonathan Pillow
This lesson is part 2 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:50:31
Speaker: : Jonathan Pillow
Course:
This lesson gives an introduction to simple spiking neuron models.
Difficulty level: Beginner
Duration: 48 Slides
Speaker: : Zubin Bhuyan
Course:
This lecture covers an Introduction to neuron anatomy and signaling, as well as different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
Course:
This lecture describes forms of plasticity on many levels: short-term, long-term, metaplasticity, and structural plasticity. Included in this lecture are also examples related to modelling of biochemical networks.
Difficulty level: Beginner
Duration: 1:11:29
Speaker: : Upi Bhalla
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Provenance (2)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (6)
- Assembly 2021 (29)
- Brain-hardware interfaces (13)
- Clinical neuroscience (17)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(45)
- Neuroscience (9)
- Cognitive Science (7)
- Cell signaling (3)
- Brain networks (4)
- Glia (1)
- Electrophysiology (16)
- Learning and memory (3)
- Neuroanatomy (17)
- Neurobiology (7)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (4)
- Neurophysiology (22)
- Neuropharmacology (2)
- Synaptic plasticity (2)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(15)
- (-) Computational neuroscience (195)
- Statistics (2)
- Computer Science (15)
- Genomics (26)
- Data science
(24)
- Open science (56)
- Project management (7)
- Education (3)
- Publishing (4)
- Neuroethics (37)