Difficulty level
This lecture covers the NIDM data format within BIDS to make your datasets more searchable, and how to optimize your dataset searches.
Difficulty level: Beginner
Duration: 12:33
Speaker: : David Keator
This lecture covers positron emission tomography (PET) imaging and the Brain Imaging Data Structure (BIDS), and how they work together within the PET-BIDS standard to make neuroscience more open and FAIR.
Difficulty level: Beginner
Duration: 12:06
Speaker: : Melanie Ganz
This lecture discusses how to standardize electrophysiology data organization to move towards being more FAIR.
Difficulty level: Beginner
Duration: 15:51
Speaker: : Sylvain Takerkart
Hierarchical Event Descriptors (HED) fill a major gap in the neuroinformatics standards toolkit, namely the specification of the nature(s) of events and time-limited conditions recorded as having occurred during time series recordings (EEG, MEG, iEEG, fMRI, etc.). Here, the HED Working Group presents an online INCF workshop on the need for, structure of, tools for, and use of HED annotation to prepare neuroimaging time series data for storing, sharing, and advanced analysis.
Difficulty level: Beginner
Duration: 03:37:42
Speaker: :
In this lesson, attendees will learn about the data structure standards, specifically the Brain Imaging Data Structure (BIDS), an INCF-endorsed standard for organizing, annotating, and describing data collected during neuroimaging experiments.
Difficulty level: Beginner
Duration: 21:56
Speaker: : Michael Schirner
This lecture covers a lot of post-war developments in the science of the mind, focusing first on the cognitive revolution, and concluding with living machines.
Difficulty level: Beginner
Duration: 2:24:35
Speaker: : Paul F.M.J. Verschure
This brief talk goes into work being done at The Alan Turing Institute to solve real-world challenges and democratize computer vision methods to support interdisciplinary and international researchers.
Difficulty level: Beginner
Duration: 7:10
Speaker: : Alden Connor & Beatriz Costa Gomes
This lesson aims to define computational neuroscience in general terms, while providing specific examples of highly successful computational neuroscience projects.
Difficulty level: Beginner
Duration: 59:21
Speaker: : Alla Borisyuk
Course:
This lecture gives an introduction to simulation, models, and the neural simulation tool NEST.
Difficulty level: Beginner
Duration: 1:48:18
Speaker: : Marc-Oliver Gewaltig
Course:
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lesson discuses forms of neural plasticity on many levels, including short-term, long-term, metaplasticity, and structural plasticity. During the lesson you will also be presented with examples related to the modelling of biochemical networks.
Difficulty level: Beginner
Duration: 1:11:29
Speaker: : Upi Bhalla
This lesson provides an introduction to modelling of chemical computation in the brain.
Difficulty level: Beginner
Duration: 1:00:11
Speaker: : Upi Bhalla
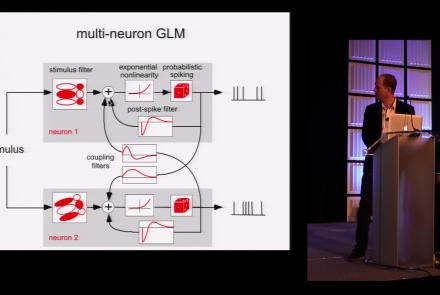
This lesson is part 1 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:45:48
Speaker: : Jonathan Pillow
This lesson is part 2 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:50:31
Speaker: : Jonathan Pillow
Course:
This lesson gives an introduction to simple spiking neuron models.
Difficulty level: Beginner
Duration: 48 Slides
Speaker: : Zubin Bhuyan
Course:
This lecture covers an Introduction to neuron anatomy and signaling, as well as different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
Course:
This lecture describes forms of plasticity on many levels: short-term, long-term, metaplasticity, and structural plasticity. Included in this lecture are also examples related to modelling of biochemical networks.
Difficulty level: Beginner
Duration: 1:11:29
Speaker: : Upi Bhalla
Course:
This lesson provides an introduction to modelling of chemical computation in the brain.
Difficulty level: Beginner
Duration: 1:00:11
Speaker: : Upi Bhalla
This lesson provides an introduction to the role of models in theoretical neuroscience, particularly focusing on David Marr's work on levels of description/analysis of the brain as a complex system: computation, algorithm & representation, and implementation.
Difficulty level: Beginner
Duration: 19:26
Speaker: : Jakob Macke
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Provenance (2)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (6)
- Assembly 2021 (29)
- Brain-hardware interfaces (13)
- Clinical neuroscience (17)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(45)
- Neuroscience (9)
- Cognitive Science (7)
- Cell signaling (3)
- Brain networks (4)
- Glia (1)
- Electrophysiology (16)
- Learning and memory (3)
- Neuroanatomy (17)
- Neurobiology (7)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (4)
- Neurophysiology (22)
- Neuropharmacology (2)
- Synaptic plasticity (2)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(15)
- (-) Computational neuroscience (195)
- Statistics (2)
- Computer Science (15)
- Genomics (26)
- Data science
(24)
- Open science (56)
- Project management (7)
- Education (3)
- Publishing (4)
- Neuroethics (37)