Lesson type
Difficulty level
This lecture covers the NIDM data format within BIDS to make your datasets more searchable, and how to optimize your dataset searches.
Difficulty level: Beginner
Duration: 12:33
Speaker: : David Keator
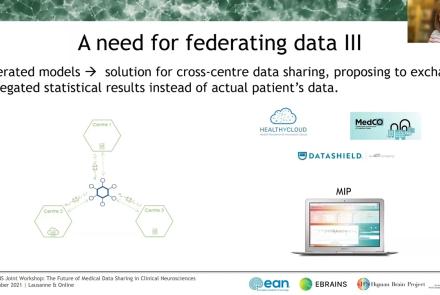
In this session the Medical Informatics Platform (MIP) federated analytics is presented. The current and future analytical tools implemented in the MIP will be detailed along with the constructs, tools, processes, and restrictions that formulate the solution provided. MIP is a platform providing advanced federated analytics for diagnosis and research in clinical neuroscience research. It is targeting clinicians, clinical scientists and clinical data scientists. It is designed to help adopt advanced analytics, explore harmonized medical data of neuroimaging, neurophysiological and medical records as well as research cohort datasets, without transferring original clinical data. It can be perceived as a virtual database that seamlessly presents aggregated data from distributed sources, provides access and analyze imaging and clinical data, securely stored in hospitals, research archives and public databases. It leverages and re-uses decentralized patient data and research cohort datasets, without transferring original data. Integrated statistical analysis tools and machine learning algorithms are exposed over harmonized, federated medical data.
Difficulty level: Intermediate
Duration: 15:05
Speaker: : Giorgos Papanikos
The Medical Informatics Platform (MIP) is a platform providing federated analytics for diagnosis and research in clinical neuroscience research. The federated analytics is possible thanks to a distributed engine that executes computations and transfers information between the members of the federation (hospital nodes). In this talk the speaker will describe the process of designing and implementing new analytical tools, i.e. statistical and machine learning algorithms. Mr. Sakellariou will further describe the environment in which these federated algorithms run, the challenges and the available tools, the principles that guide its design and the followed general methodology for each new algorithm. One of the most important challenges which are faced is to design these tools in a way that does not compromise the privacy of the clinical data involved. The speaker will show how to address the main questions when designing such algorithms: how to decompose and distribute the computations and what kind of information to exchange between nodes, in order to comply with the privacy constraint mentioned above. Finally, also the subject of validating these federated algorithms will be briefly touched.
Difficulty level: Intermediate
Duration: 20:26
Speaker: : Jason Skellariou
The Medical Informatics Platform (MIP) Dementia had been installed in several memory clinics across Europe allowing them to federate their real-world databases. Research open access databases had also been integrated such as ADNI (Alzheimer’s Dementia Neuroimaging Initiative), reaching a cumulative case load of more than 5,000 patients (major cognitive disorder due to Alzheimer’s disease, other major cognitive disorder, minor cognitive disorder, controls). The statistic and machine learning tools implemented in the MIP allowed researchers to conduct easily federated analyses among Italian memory clinics (Redolfi et al. 2020) and also across borders between the French (Lille), the Swiss (Lausanne) and the Italian (Brescia) datasets.
Difficulty level: Intermediate
Duration: 16:44
Speaker: : Mélanie Leroy
This talks presents ethics requirements of the Medical Informatics Platform, a data sharing platform for medical data using data federation mechanisms. The talk presents how the Medical Informatics Platform (MIP) works and which ethical requirements need to be considered when working with federated data.
Difficulty level: Intermediate
Duration: 16:25
Speaker: : Erika Borcel
This lecture talks about the usage of knowledge graphs in hospitals and related challenges of semantic interoperability.
Difficulty level: Intermediate
Duration: 24:32
Speaker: : Cristophe Gaudet-Blavignac
This lecture discusses risk-based anonymization approaches for medical research.
Difficulty level: Intermediate
Duration: 15:43
Speaker: : Fabian Prasser
This lesson introduces concepts and practices surrounding reference atlases for the mouse and rat brains. Additionally, this lesson provides discussion around examples of data systems employed to organize neuroscience data collections in the context of reference atlases as well as analytical workflows applied to the data.
Difficulty level: Beginner
Duration: 03:04:29
Speaker: :
This talk covers EBRAINS, an open research infrastructure that gathers data, tools and computing facilities for brain-related research, built with interoperability at the core.
Difficulty level: Beginner
Duration: 8:22
Speaker: : Petra Ritter
This lesson provides an introduction to the European open research infrastructure EBRAINS and its digital brain atlas resources.
Difficulty level: Beginner
Duration: 27:45
Speaker: : Trygve Leergard
This lesson contains both a lecture and a tutorial component. The lecture (0:00-20:03 of YouTube video) discusses both the need for intersectional approaches in healthcare as well as the impact of neglecting intersectionality in patient populations. The lecture is followed by a practical tutorial in both Python and R on how to assess intersectional bias in datasets. Links to relevant code and data are found below.
Difficulty level: Beginner
Duration: 52:26
Course:
This is an introductory lecture on whole-brain modelling, delving into the various spatial scales of neuroscience, neural population models, and whole-brain modelling. Additionally, the clinical applications of building and testing such models are characterized.
Difficulty level: Intermediate
Duration: 1:24:44
Speaker: : John Griffiths
This is a tutorial on designing a Bayesian inference model to map belief trajectories, with emphasis on gaining familiarity with Hierarchical Gaussian Filters (HGFs).
This lesson corresponds to slides 65-90 of the PDF below.
Difficulty level: Intermediate
Duration: 1:15:04
Speaker: : Daniel Hauke
This lecture discusses what defines an integrative approach regarding research and methods, including various study designs and models which are appropriate choices when attempting to bridge data domains; a necessity when whole-person modelling.
Difficulty level: Beginner
Duration: 1:28:14
Speaker: : Dan Felsky
Similarity Network Fusion (SNF) is a computational method for data integration across various kinds of measurements, aimed at taking advantage of the common as well as complementary information in different data types. This workshop walks participants through running SNF on EEG and genomic data using RStudio.
Difficulty level: Intermediate
Duration: 1:21:38
Speaker: : Dan Felsky
In this lesson, you will learn about one particular aspect of decision making: reaction times. In other words, how long does it take to take a decision based on a stream of information arriving continuously over time?
Difficulty level: Intermediate
Duration: 6:01
Speaker: : Dan Goodman
Course:
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lecture covers an Introduction to neuron anatomy and signaling, and different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
Course:
This lecture covers an Introduction to neuron anatomy and signaling, as well as different types of models, including the Hodgkin-Huxley model.
Difficulty level: Beginner
Duration: 1:23:01
Speaker: : Gaute Einevoll
This lecture describes non-spiking simple neuron models used in artificial neural networks and machine learning.
Difficulty level: Beginner
Duration: 8:23
Speaker: : Geoffrey Hinton
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)