Lesson type
Difficulty level
Course:
After introducing the local epileptor model in the previous two videos, we will now use it in a large-scale brain simulation. We again focus on the paper The Virtual Epileptic Patient: Individualized whole-brain models of epilepsy spread. Two simulations with different epileptogenicity across the network are visualized to show the difference in seizure spread across the cortex.
Difficulty level: Beginner
Duration: 6:36
Speaker: : Paul Triebkorn
Course:
This lecture gives an overview on the article Individual brain structure and modelling predict seizure propagation, in which 15 subjects with epilepsy were modelled to predict individual epileptogenic zones. With the TVB GUI we will model seizure spread and the effect of lesioning the connectome. The impact of cutting edges in the network on seizure spreading will be visualized.
Difficulty level: Beginner
Duration: 9:39
Speaker: : Paul Triebkorn
This lecture briefly introduces The Virtual Brain (TVB), a multi-scale, multi-modal neuroinformatics platform for full brain network simulations using biologically realistic connectivity, as well as its potential neuroscience applications (e.g., epilepsy cases).
Difficulty level: Beginner
Duration: 8:53
Speaker: : Petra Ritter
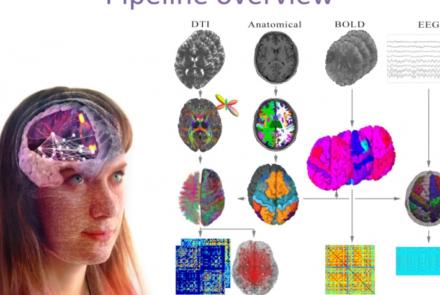
This presentation accompanies the paper entitled: An automated pipeline for constructing personalized virtual brains from multimodal neuroimaging data (see link below to download publication).
Difficulty level: Beginner
Duration: 4:56
Course:
In this lecture, attendees will learn how Mutant Mouse Resource and Research Center (MMRRC) archives, cryopreserves, and distributes scientifically valuable genetically engineered mouse strains and mouse ES cell lines for the genetics and biomedical research community.
Difficulty level: Beginner
Duration: 43:38
Speaker: : Kent Lloyd
Course:
This lesson provides a brief overview of the Python programming language, with an emphasis on tools relevant to data scientists.
Difficulty level: Beginner
Duration: 1:16:36
Speaker: : Tal Yarkoni
This lecture on model types introduces the advantages of modeling, provide examples of different model types, and explain what modeling is all about.
Difficulty level: Beginner
Duration: 27:48
Speaker: : Gunnar Blohm
Course:
This lecture focuses on how to get from a scientific question to a model using concrete examples. We will present a 10-step practical guide on how to succeed in modeling. This lecture contains links to 2 tutorials, lecture/tutorial slides, suggested reading list, and 3 recorded Q&A sessions.
Difficulty level: Beginner
Duration: 29:52
Speaker: : Megan Peters
Course:
This lecture formalizes modeling as a decision process that is constrained by a precise problem statement and specific model goals. We provide real-life examples on how model building is usually less linear than presented in Modeling Practice I.
Difficulty level: Beginner
Duration: 22:51
Speaker: : Gunnar Blohm
Course:
This lecture focuses on the purpose of model fitting, approaches to model fitting, model fitting for linear models, and how to assess the quality and compare model fits. We will present a 10-step practical guide on how to succeed in modeling.
Difficulty level: Beginner
Duration: 26:46
Speaker: : Jan Drugowitsch
Course:
This lecture summarizes the concepts introduced in Model Fitting I and adds two additional concepts: 1) MLE is a frequentist way of looking at the data and the model, with its own limitations. 2) Side-by-side comparisons of bootstrapping and cross-validation.
Difficulty level: Beginner
Duration: 38.17
Speaker: : Kunlin Wei
This lecture provides an overview of the generalized linear models (GLM) course, originally a part of the Neuromatch Academy (NMA), an interactive online summer school held in 2020. NMA provided participants with experiences spanning from hands-on modeling experience to meta-science interpretation skills across just about everything that could reasonably be included in the label "computational neuroscience".
Difficulty level: Beginner
Duration: 33:58
Speaker: : Cristina Savin
This lecture further develops the concepts introduced in Machine Learning I. This lecture is part of the Neuromatch Academy (NMA), an interactive online computational neuroscience summer school held in 2020.
Difficulty level: Beginner
Duration: 29:30
Speaker: : I. Memming Park
This lesson provides an overview of the process of developing the TVB-NEST co-simulation on the EBRAINS infrastructure, and its use cases.
Difficulty level: Beginner
Duration: 25:14
Speaker: : Denis Perdikis
Course:
This lecture introduces the core concepts of dimensionality reduction.
Difficulty level: Beginner
Duration: 31:43
Speaker: : Byron Yu
Course:
This lecture covers the application of dimensionality reduction applied to multi-dimensional neural recordings using brain-computer interfaces with simultaneous spike recordings.
Difficulty level: Beginner
Duration: 30:15
Speaker: : Byron Yu
This is a tutorial covering Generalized Linear Models (GLMs), which are a fundamental framework for supervised learning. In this tutorial, the objective is to model a retinal ganglion cell spike train by fitting a temporal receptive field: first with a Linear-Gaussian GLM (also known as ordinary least-squares regression model) and then with a Poisson GLM (aka "Linear-Nonlinear-Poisson" model). The data you will be using was published by Uzzell & Chichilnisky 2004.
Difficulty level: Beginner
Duration: 8:09
Speaker: : Anqi Wu
Course:
This tutorial covers multivariate data can be represented in different orthonormal bases.
Difficulty level: Beginner
Duration: 4:48
Speaker: : Alex Cayco Gajic
Course:
This tutorial covers how to perform principal component analysis (PCA) by projecting the data onto the eigenvectors of its covariance matrix.
To quickly refresh your knowledge of eigenvalues and eigenvectors, you can watch this short video (4 minutes) for a geometrical explanation. For a deeper understanding, this in-depth video (17 minutes) provides an excellent basis and is beautifully illustrated.
Difficulty level: Beginner
Duration: 6:33
Speaker: : Alex Cayco Gajic
Course:
This tutorial covers how to apply principal component analysis (PCA) for dimensionality reduction, using a classic dataset that is often used to benchmark machine learning algorithms: MNIST. We'll also learn how to use PCA for reconstruction and denoising.
You can learn more about MNIST dataset here.
Difficulty level: Beginner
Duration: 5:35
Speaker: : Alex Cayco Gajic
Topics
- Philosophy of Science (5)
- Artificial Intelligence (4)
- Animal models (3)
- Assembly 2021 (27)
- Brain-hardware interfaces (1)
- Clinical neuroscience (11)
- International Brain Initiative (2)
- Repositories and science gateways (5)
- Resources (6)
- General neuroscience
(9)
- Phenome (1)
- General neuroinformatics (2)
- (-) Computational neuroscience (67)
- Statistics (1)
- (-) Computer Science (5)
- Genomics (22)
- Data science (17)
- Open science (29)
- Project management (3)
- Education (2)
- Publishing (1)
- Neuroethics (6)