Lesson type
Difficulty level
Course:
This lesson describes the principles underlying functional magnetic resonance imaging (fMRI), diffusion-weighted imaging (DWI), tractography, and parcellation. These tools and concepts are explained in a broader context of neural connectivity and mental health.
Difficulty level: Intermediate
Duration: 1:47:22
Speaker: : Erin Dickie and John Griffiths
Course:
This tutorial introduces pipelines and methods to compute brain connectomes from fMRI data. With corresponding code and repositories, participants can follow along and learn how to programmatically preprocess, curate, and analyze functional and structural brain data to produce connectivity matrices.
Difficulty level: Intermediate
Duration: 1:39:04
Speaker: : Erin Dickie and John Griffiths
This lesson introduces the practical exercises which accompany the previous lessons on animal and human connectomes in the brain and nervous system.
Difficulty level: Intermediate
Duration: 4:10
Speaker: : Dan Goodman
Course:
This lecture and tutorial focuses on measuring human functional brain networks, as well as how to account for inherent variability within those networks.
Difficulty level: Intermediate
Duration: 50:44
Speaker: : Caterina Gratton
Course:
This lecture presents an overview of functional brain parcellations, as well as a set of tutorials on bootstrap agregation of stable clusters (BASC) for fMRI brain parcellation.
Difficulty level: Advanced
Duration: 50:28
Speaker: : Pierre Bellec
Course:
This presentation by the OHBM OpenScienceSIG covers common scenarios where Git can be extremely valuable. The essentials covered include cloning a repository and keeping it up to date, how to create and use your own repository, and how to contribute to other projects via forking and pull requests.
Difficulty level: Beginner
Duration: 51:55
Speaker: : Saskia Bollmann, Steffen Bollmann
Course:
DataLad is a versatile data management and data publication multi-tool. In this session, you can learn the basic concepts and commands for version control and reproducible data analysis. You’ll get to see, create, and install DataLad datasets of many shapes and sizes, master local version workflows and provenance-captured analysis-execution, and you will get ideas for your next data analysis project.
Difficulty level: Beginner
Duration: 01:29:08
Speaker: : Adina Wagner
Course:
Overview of the content for Day 1 of this course.
Difficulty level: Beginner
Duration: 00:01:59
Speaker: : Tristan Shuman
Course:
Overview of Day 2 of this course.
Difficulty level: Beginner
Duration: 00:03:28
Speaker: : Tristan Shuman
Course:
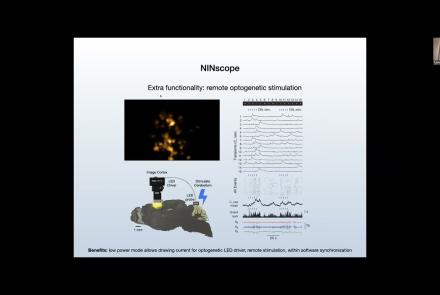
Best practices: the tips and tricks on how to get your Miniscope to work and how to get your experiments off the ground.
Difficulty level: Beginner
Duration: 00:53:34
Course:
This talk compares various sensors and resolutions for in vivo neural recordings.
Difficulty level: Beginner
Duration: 00:24:03
Speaker: : Susie Feng, Zach Pennington, Caleb Kemere
Course:
This talk delves into challenges and opportunities of Miniscope design, seeking the optimal balance between scale and function.
Difficulty level: Beginner
Duration: 00:21:51
Speaker: : Susie Feng, Zach Pennington, Tycho Hoogland
Course:
Attendees of this talk will learn aobut computational imaging systems and associated pipelines, as well as open-source software solutions supporting miniscope use.
Difficulty level: Beginner
Duration: 00:17:56
Speaker: : Susie Feng, Zach Pennington, Laura Waller
Course:
This talk covers the present state and future directions of calcium imaging data analysis, particularly in the context of one-photon vs two-photon approaches.
Difficulty level: Beginner
Duration: 00:21:06
Course:
In this talk, results from rodent experimentation using in vivo imaging are presented, demonstrating how the monitoring of neural ensembles may reveal patterns of learning during spatial tasks.
Difficulty level: Beginner
Duration: 00:19:43
Speaker: : Susie Feng, Zach Pennington, William Mau
Course:
How to start processing the raw imaging data generated with a Miniscope, including developing a usable pipeline and demoing the Minion pipeline.
Difficulty level: Beginner
Duration: 00:57:26
Speaker: : Daniel Aharoni, Phil Dong
Course:
The direction of miniature microscopes, including both MetaCell and other groups.
Difficulty level: Beginner
Duration: 00:49:16
Speaker: : Daniel Aharoni, Frederico N Sangiuliano
Course:
Overview of the content for Day 2 of this course.
Difficulty level: Beginner
Duration: 00:11:01
Speaker: : Tristan Shuman
Course:
Summary and closing remarks for this three-day course.
Difficulty level: Beginner
Duration: 00:04:56
Speaker: : Stephen Larson
Course:
This hands-on tutorial explains how to run your own Minion session in the MetaCell cloud using jupityr notebooks.
Difficulty level: Beginner
Duration: 01:28:03
Speaker: : Daniel Aharoni, Phil Dong
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)