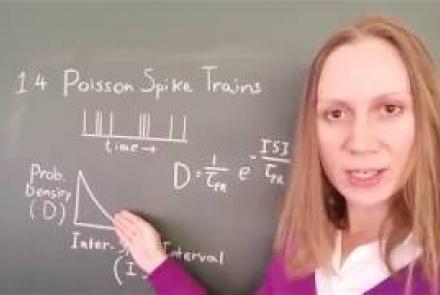Lesson type
Difficulty level
This lesson describes spike timing-dependent plasticity (STDP), a biological process that adjusts the strength of connections between neurons in the brain, and how one can implement or mimic this process in a computational model. You will also find links for practical exercises at the bottom of this page.
Difficulty level: Intermediate
Duration: 12:50
Speaker: : Dan Goodman
This lesson provides a brief introduction to the Computational Modeling of Neuronal Plasticity.
Difficulty level: Intermediate
Duration: 0:40
Speaker: : Florence I. Kleberg
In this lesson, you will be introducted to a type of neuronal model known as the leaky integrate-and-fire (LIF) model.
Difficulty level: Intermediate
Duration: 1:23
Speaker: : Florence I. Kleberg
This lesson goes over various potential inputs to neuronal synapses, loci of neural communication.
Difficulty level: Intermediate
Duration: 1:20
Speaker: : Florence I. Kleberg
This lesson describes the how and why behind implementing integration time steps as part of a neuronal model.
Difficulty level: Intermediate
Duration: 1:08
Speaker: : Florence I. Kleberg
In this lesson, you will learn about neural spike trains which can be characterized as having a Poisson distribution.
Difficulty level: Intermediate
Duration: 1:18
Speaker: : Florence I. Kleberg
This lesson covers spike-rate adaptation, the process by which a neuron's firing pattern decays to a low, steady-state frequency during the sustained encoding of a stimulus.
Difficulty level: Intermediate
Duration: 1:26
Speaker: : Florence I. Kleberg
This lesson provides a brief explanation of how to implement a neuron's refractory period in a computational model.
Difficulty level: Intermediate
Duration: 0:42
Speaker: : Florence I. Kleberg
In this lesson, you will learn a computational description of the process which tunes neuronal connectivity strength, spike-timing-dependent plasticity (STDP).
Difficulty level: Intermediate
Duration: 2:40
Speaker: : Florence I. Kleberg
This lesson reviews theoretical and mathematical descriptions of correlated spike trains.
Difficulty level: Intermediate
Duration: 2:54
Speaker: : Florence I. Kleberg
This lesson investigates the effect of correlated spike trains on spike-timing dependent plasticity (STDP).
Difficulty level: Intermediate
Duration: 1:43
Speaker: : Florence I. Kleberg
This lesson goes over synaptic normalisation, the homeostatic process by which groups of weighted inputs scale up or down their biases.
Difficulty level: Intermediate
Duration: 2:58
Speaker: : Florence I. Kleberg
In this lesson, you will learn about the intrinsic plasticity of single neurons.
Difficulty level: Intermediate
Duration: 2:08
Speaker: : Florence I. Kleberg
This lesson covers short-term facilitation, a process whereby a neuron's synaptic transmission is enhanced for a short (sub-second) period.
Difficulty level: Intermediate
Duration: 1:58
Speaker: : Florence I. Kleberg
This lesson describes short-term depression, a reduction of synaptic information transfer between neurons.
Difficulty level: Intermediate
Duration: 1:40
Speaker: : Florence I. Kleberg
This lesson briefly wraps up the course on Computational Modeling of Neuronal Plasticity.
Difficulty level: Intermediate
Duration: 0:37
Speaker: : Florence I. Kleberg
Course:
The goal of computational modeling in behavioral and psychological science is using mathematical models to characterize behavioral (or neural) data. Over the past decade, this practice has revolutionized social psychological science (and neuroscience) by allowing researchers to formalize theories as constrained mathematical models and test specific hypotheses to explain unobservable aspects of complex social cognitive processes and behaviors. This course is composed of 4 modules in the format of Jupyter Notebooks. This course comprises lecture-based, discussion-based, and lab-based instruction. At least one-third of class sessions will be hands-on. We will discuss relevant book chapters and journal articles, and work with simulated and real data using the Python programming language (no prior programming experience necessary) as we survey some selected areas of research at the intersection of computational modeling and social behavior. These selected topics will span a broad set of social psychological abilities including (1) learning from and for others, (2) learning about others, and (3) social influence on decision-making and mental states. Rhoads, S. A. & Gan, L. (2022). Computational models of human social behavior and neuroscience - An open educational course and Jupyter Book to advance computational training. Journal of Open Source Education, 5(47), 146. https://doi.org/10.21105/jose.00146
Difficulty level: Intermediate
Duration:
Speaker: :
This is the first of two workshops on reproducibility in science, during which participants are introduced to concepts of FAIR and open science. After discussing the definition of and need for FAIR science, participants are walked through tutorials on installing and using Github and Docker, the powerful, open-source tools for versioning and publishing code and software, respectively.
Difficulty level: Intermediate
Duration: 1:20:58
Speaker: : Erin Dickie and Sejal Patel
This lesson contains both a lecture and a tutorial component. The lecture (0:00-20:03 of YouTube video) discusses both the need for intersectional approaches in healthcare as well as the impact of neglecting intersectionality in patient populations. The lecture is followed by a practical tutorial in both Python and R on how to assess intersectional bias in datasets. Links to relevant code and data are found below.
Difficulty level: Beginner
Duration: 52:26
This is a hands-on tutorial on PLINK, the open source whole genome association analysis toolset. The aims of this tutorial are to teach users how to perform basic quality control on genetic datasets, as well as to identify and understand GWAS summary statistics.
Difficulty level: Intermediate
Duration: 1:27:18
Speaker: : Dan Felsky
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- (-) Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- (-) Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)