Lesson type
Difficulty level
This lightning talk describes an automated pipline for positron emission tomography (PET) data.
Difficulty level: Intermediate
Duration: 7:27
Speaker: : Soodeh Moallemian
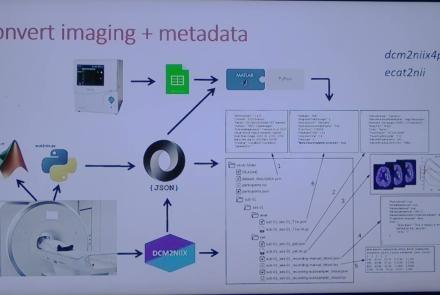
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 41:04
Speaker: : Martin Nørgaard
This session dives into practical PET tooling on BIDS data—showing how to run motion correction, register PET↔MRI, extract time–activity curves, and generate standardized PET-BIDS derivatives with clear QC reports. It introduces modular BIDS Apps (head-motion correction, TAC extraction), a full pipeline (PETPrep), and a PET/MRI defacer, with guidance on parameters, outputs, provenance, and why Dockerized containers are the reliable way to run them at scale.
Difficulty level: Intermediate
Duration: 1:05:38
Speaker: : Martin Nørgaard
This session introduces two PET quantification tools—bloodstream for processing arterial blood data and kinfitr for kinetic modeling and quantification—built to work with BIDS/BIDS-derivatives and containers. Bloodstream fuses autosampler and manual measurements (whole blood, plasma, parent fraction) using interpolation or fitted models (incl. hierarchical GAMs) to produce a clean arterial input function (AIF) and whole-blood curves with rich QC reports ready. TAC data (e.g., from PETPrep) and blood (e.g., from bloodstream) can be ingested using kinfitr to run reproducible, GUI-driven analyses: define combined ROIs, calculate weighting factors, estimate blood–tissue delay, choose and chain models (e.g., 2TCM → 1TCM with parameter inheritance), and export parameters/diagnostics. Both are available as Docker apps; workflows emphasize configuration files, reports, and standard outputs to support transparency and reuse.
Difficulty level: Intermediate
Duration: 1:20:56
Speaker: : Granville Matheson
This lecture covers positron emission tomography (PET) imaging and the Brain Imaging Data Structure (BIDS), and how they work together within the PET-BIDS standard to make neuroscience more open and FAIR.
Difficulty level: Beginner
Duration: 12:06
Speaker: : Melanie Ganz
Course:
This module covers many of the types of non-invasive neurotech and neuroimaging devices including electroencephalography (EEG), electromyography (EMG), electroneurography (ENG), magnetoencephalography (MEG), and more.
Difficulty level: Beginner
Duration: 13:36
Speaker: : Harrison Canning
This lecture covers the history of behaviorism and the ultimate challenge to behaviorism.
Difficulty level: Beginner
Duration: 1:19:08
Speaker: : Paul F.M.J. Verschure
In this lesson, you will learn how to utilize various features and tools included in the EBRAINS platform, particularly focusing on rodent brain atlases and how to incorporate them into your analyses.
Difficulty level: Beginner
Duration: 15:48
Speaker: : Sharon Yates
This talk describes how to use DataLad for your data management and curation techniques when dealing with animal datasets, which often contain several disparate types of data, including MRI, microscopy, histology, electrocorticography, and behavioral measurements.
Difficulty level: Beginner
Duration: 3:35
Speaker: : Aref Kalantari Sarcheshmeh
In this short talk you will learn about The Neural System Laboratory, which aims to develop and implement new technologies for analysis of brain architecture, connectivity, and brain-wide gene and molecular level organization.
Difficulty level: Beginner
Duration: 8:38
Speaker: : Trygve Leergard
In this lesson, you will learn about the connectome, the collective system of neural pathways in an organism, with a closer look at the neurons, synapses, and connections of particular species.
Difficulty level: Intermediate
Duration: 6:48
Speaker: : Marcus Ghosh
This lesson introduces the practical exercises which accompany the previous lessons on animal and human connectomes in the brain and nervous system.
Difficulty level: Intermediate
Duration: 4:10
Speaker: : Dan Goodman
Course:
In this lecture, attendees will learn how Mutant Mouse Resource and Research Center (MMRRC) archives, cryopreserves, and distributes scientifically valuable genetically engineered mouse strains and mouse ES cell lines for the genetics and biomedical research community.
Difficulty level: Beginner
Duration: 43:38
Speaker: : Kent Lloyd
This lecture discusses how to standardize electrophysiology data organization to move towards being more FAIR.
Difficulty level: Beginner
Duration: 15:51
Speaker: : Sylvain Takerkart
While the previous lesson in the Neuro4ML course dealt with the mechanisms involved in individual synapses, this lesson discusses how synapses and their neurons' firing patterns may change over time.
Difficulty level: Intermediate
Duration: 4:48
Speaker: : Marcus Ghosh
In this lesson, you will learn about how machine learners and computational neuroscientists design and build models of neuronal synapses.
Difficulty level: Intermediate
Duration: 8:59
Speaker: : Dan Goodman
Course:
How does the brain learn? This lecture discusses the roles of development and adult plasticity in shaping functional connectivity.
Difficulty level: Beginner
Duration: 1:08:45
Speaker: : Clay Reid
This lesson goes into the mechanisms behind changes in synaptic function created by learning.
Difficulty level: Beginner
Duration: 27:07
Speaker: : Carl Petersen
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- (-) Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- (-) Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)