Lesson type
Difficulty level
This talk enumerates the challenges regarding data accessibility and reusability inherent in the current scientific publication system, and discusses novel approaches to these challenges, such as the EBRAINS Live Papers platform.
Difficulty level: Beginner
Duration: 18:08
Speaker: : Andrew Davison
This brief video gives an introduction to the eighth session of INCF's Neuroinformatics Assembly 2023, focusing on FAIR data and the role of academic journals.
Difficulty level: Beginner
Duration: 5:57
Speaker: : Jan G. Bjaalie
This talk gives an overview of the perspectives and FAIR-aligned policies of the academic journal Public Library of Science, better known as PLOS. This journal is a nonprofit, open access publisher empowering researchers to accelerate progress in science.
Difficulty level: Beginner
Duration: 11:53
Speaker: : Iain Hrynaszkiewicz
This talk highlights a set of platform technologies, software, and data collections that close and shorten the feedback cycle in research.
Difficulty level: Beginner
Duration: 57:52
Speaker: : Satrajit Ghosh
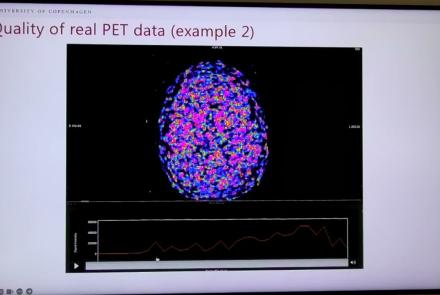
This lightning talk describes an automated pipline for positron emission tomography (PET) data.
Difficulty level: Intermediate
Duration: 7:27
Speaker: : Soodeh Moallemian
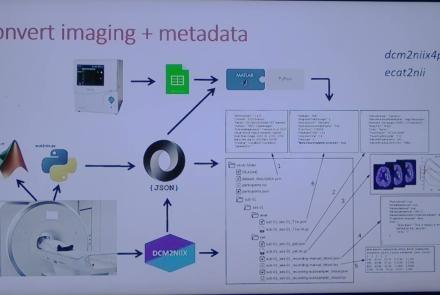
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 41:04
Speaker: : Martin Nørgaard
This session dives into practical PET tooling on BIDS data—showing how to run motion correction, register PET↔MRI, extract time–activity curves, and generate standardized PET-BIDS derivatives with clear QC reports. It introduces modular BIDS Apps (head-motion correction, TAC extraction), a full pipeline (PETPrep), and a PET/MRI defacer, with guidance on parameters, outputs, provenance, and why Dockerized containers are the reliable way to run them at scale.
Difficulty level: Intermediate
Duration: 1:05:38
Speaker: : Martin Nørgaard
This session introduces two PET quantification tools—bloodstream for processing arterial blood data and kinfitr for kinetic modeling and quantification—built to work with BIDS/BIDS-derivatives and containers. Bloodstream fuses autosampler and manual measurements (whole blood, plasma, parent fraction) using interpolation or fitted models (incl. hierarchical GAMs) to produce a clean arterial input function (AIF) and whole-blood curves with rich QC reports ready. TAC data (e.g., from PETPrep) and blood (e.g., from bloodstream) can be ingested using kinfitr to run reproducible, GUI-driven analyses: define combined ROIs, calculate weighting factors, estimate blood–tissue delay, choose and chain models (e.g., 2TCM → 1TCM with parameter inheritance), and export parameters/diagnostics. Both are available as Docker apps; workflows emphasize configuration files, reports, and standard outputs to support transparency and reuse.
Difficulty level: Intermediate
Duration: 1:20:56
Speaker: : Granville Matheson
This lecture covers positron emission tomography (PET) imaging and the Brain Imaging Data Structure (BIDS), and how they work together within the PET-BIDS standard to make neuroscience more open and FAIR.
Difficulty level: Beginner
Duration: 12:06
Speaker: : Melanie Ganz
Course:
This module covers many of the types of non-invasive neurotech and neuroimaging devices including electroencephalography (EEG), electromyography (EMG), electroneurography (ENG), magnetoencephalography (MEG), and more.
Difficulty level: Beginner
Duration: 13:36
Speaker: : Harrison Canning
Course:
This lecture covers computational principles that growth cones employ to detect and respond to environmental chemotactic gradients, focusing particularly on growth-cone shape dynamics.
Difficulty level: Intermediate
Duration: 26:12
Speaker: : Geoff Goodhill
Course:
In this lecture you will learn that in developing mouse somatosensory cortex, endogenous Btbd3 translocate to the cell nucleus in response to neuronal activity and oriente primary dendrites toward active axons in the barrel hollow.
Difficulty level: Intermediate
Duration: 27:32
Speaker: : Tomomi Shimogori
Course:
In this presentation, the speaker describes some of their recent efforts to characterize the transcriptome of the developing human brain, and and introduction to the BrainSpan project.
Difficulty level: Intermediate
Duration: 30:45
Speaker: : Nenad Sestan
Course:
How does the brain learn? This lecture discusses the roles of development and adult plasticity in shaping functional connectivity.
Difficulty level: Beginner
Duration: 1:08:45
Speaker: : Clay Reid
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- (-) Publishing (4)
- Neuroethics (42)