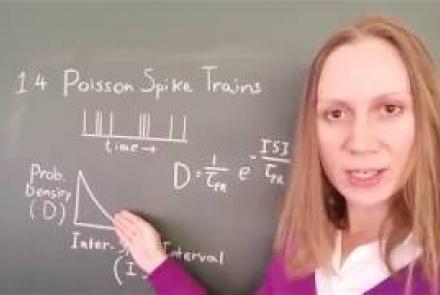Difficulty level
This lesson describes spike timing-dependent plasticity (STDP), a biological process that adjusts the strength of connections between neurons in the brain, and how one can implement or mimic this process in a computational model. You will also find links for practical exercises at the bottom of this page.
Difficulty level: Intermediate
Duration: 12:50
Speaker: : Dan Goodman
This lesson provides a brief introduction to the Computational Modeling of Neuronal Plasticity.
Difficulty level: Intermediate
Duration: 0:40
Speaker: : Florence I. Kleberg
In this lesson, you will be introducted to a type of neuronal model known as the leaky integrate-and-fire (LIF) model.
Difficulty level: Intermediate
Duration: 1:23
Speaker: : Florence I. Kleberg
This lesson goes over various potential inputs to neuronal synapses, loci of neural communication.
Difficulty level: Intermediate
Duration: 1:20
Speaker: : Florence I. Kleberg
This lesson describes the how and why behind implementing integration time steps as part of a neuronal model.
Difficulty level: Intermediate
Duration: 1:08
Speaker: : Florence I. Kleberg
In this lesson, you will learn about neural spike trains which can be characterized as having a Poisson distribution.
Difficulty level: Intermediate
Duration: 1:18
Speaker: : Florence I. Kleberg
This lesson covers spike-rate adaptation, the process by which a neuron's firing pattern decays to a low, steady-state frequency during the sustained encoding of a stimulus.
Difficulty level: Intermediate
Duration: 1:26
Speaker: : Florence I. Kleberg
This lesson provides a brief explanation of how to implement a neuron's refractory period in a computational model.
Difficulty level: Intermediate
Duration: 0:42
Speaker: : Florence I. Kleberg
In this lesson, you will learn a computational description of the process which tunes neuronal connectivity strength, spike-timing-dependent plasticity (STDP).
Difficulty level: Intermediate
Duration: 2:40
Speaker: : Florence I. Kleberg
This lesson reviews theoretical and mathematical descriptions of correlated spike trains.
Difficulty level: Intermediate
Duration: 2:54
Speaker: : Florence I. Kleberg
This lesson investigates the effect of correlated spike trains on spike-timing dependent plasticity (STDP).
Difficulty level: Intermediate
Duration: 1:43
Speaker: : Florence I. Kleberg
This lesson goes over synaptic normalisation, the homeostatic process by which groups of weighted inputs scale up or down their biases.
Difficulty level: Intermediate
Duration: 2:58
Speaker: : Florence I. Kleberg
In this lesson, you will learn about the intrinsic plasticity of single neurons.
Difficulty level: Intermediate
Duration: 2:08
Speaker: : Florence I. Kleberg
This lesson covers short-term facilitation, a process whereby a neuron's synaptic transmission is enhanced for a short (sub-second) period.
Difficulty level: Intermediate
Duration: 1:58
Speaker: : Florence I. Kleberg
This lesson describes short-term depression, a reduction of synaptic information transfer between neurons.
Difficulty level: Intermediate
Duration: 1:40
Speaker: : Florence I. Kleberg
This lesson briefly wraps up the course on Computational Modeling of Neuronal Plasticity.
Difficulty level: Intermediate
Duration: 0:37
Speaker: : Florence I. Kleberg
This lightning talk describes an automated pipline for positron emission tomography (PET) data.
Difficulty level: Intermediate
Duration: 7:27
Speaker: : Soodeh Moallemian
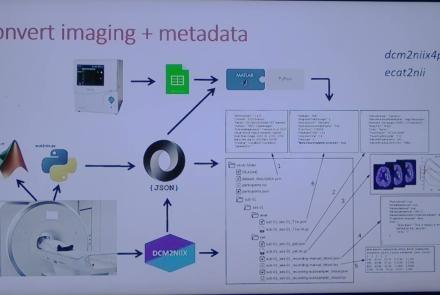
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 41:04
Speaker: : Martin Nørgaard
This session dives into practical PET tooling on BIDS data—showing how to run motion correction, register PET↔MRI, extract time–activity curves, and generate standardized PET-BIDS derivatives with clear QC reports. It introduces modular BIDS Apps (head-motion correction, TAC extraction), a full pipeline (PETPrep), and a PET/MRI defacer, with guidance on parameters, outputs, provenance, and why Dockerized containers are the reliable way to run them at scale.
Difficulty level: Intermediate
Duration: 1:05:38
Speaker: : Martin Nørgaard
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- (-) Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)