Lesson type
Difficulty level
This lesson continues with the second workshop on reproducible science, focusing on additional open source tools for researchers and data scientists, such as the R programming language for data science, as well as associated tools like RStudio and R Markdown. Additionally, users are introduced to Python and iPython notebooks, Google Colab, and are given hands-on tutorials on how to create a Binder environment, as well as various containers in Docker and Singularity.
Difficulty level: Beginner
Duration: 1:16:04
Speaker: : Erin Dickie and Sejal Patel
This talk describes the relevance and power of using brain atlases as part of one's data integration pipeline.
Difficulty level: Beginner
Duration: 15:37
Speaker: : Timo Dickscheid
In this lesson, you will learn how to understand data management plans and why data sharing is important.
Difficulty level: Beginner
Duration: 44:24
Speaker: : Jenny Muilenburg
Course:
This quick visual walkthrough presents the steps required in uploading data into a brainlife project using the graphical user interface (GUI).
Difficulty level: Beginner
Duration: 2:00
Speaker: :
Course:
This short walkthrough documents the steps needed to find a dataset in OpenNeuro, a free and open platform for sharing MRI, MEG, EEG, iEEG, ECoG, ASL, and PET data, and import it directly to a brainlife project.
Difficulty level: Beginner
Duration: 0:35
Speaker: :
Course:
This lesson describes and shows four different ways one may upload their data to brainlife.io.
Difficulty level: Beginner
Duration: 6:35
Speaker: :
Course:
This lecture covers why data sharing and other collaborative practices are important, how these practices are developed, and the challenges involved in their development and implementation.
Difficulty level: Beginner
Duration: 11:41
Speaker: : Joost Wagenaar
This lecture discusses the FAIR principles as they apply to electrophysiology data and metadata, the building blocks for community tools and standards, platforms and grassroots initiatives, and the challenges therein.
Difficulty level: Beginner
Duration: 8:11
Speaker: : Thomas Wachtler
This lecture contains an overview of electrophysiology data reuse within the EBRAINS ecosystem.
Difficulty level: Beginner
Duration: 15:57
Speaker: : Andrew Davison
This lecture contains an overview of the Distributed Archives for Neurophysiology Data Integration (DANDI) archive, its ties to FAIR and open-source, integrations with other programs, and upcoming features.
Difficulty level: Beginner
Duration: 13:34
Speaker: : Yaroslav O. Halchenko
This lesson provides a short overview of the main features of the Canadian Open Neuroscience Platform (CONP) Portal, a web interface that facilitates open science for the neuroscience community by simplifying global access to and sharing of datasets and tools. The Portal internalizes the typical cycle of a research project, beginning with data acquisition, followed by data processing with published tools, and ultimately the publication of results with a link to the original dataset.
Difficulty level: Beginner
Duration: 14:03
Speaker: : Samir Das, Tristan Glatard
This is a tutorial on how to simulate neuronal spiking in brain microcircuit models, as well as how to analyze, plot, and visualize the corresponding data.
Difficulty level: Intermediate
Duration: 1:39:50
Speaker: : Frank Mazza
Course:
This video will document the process of running an app on brainlife, from data staging to archiving of the final data outputs.
Difficulty level: Beginner
Duration: 3:43
Speaker: :
Course:
This quick video presents some of the various visualizers available on brainlife.io
Difficulty level: Beginner
Duration: 1:11
Speaker: :
Course:
This short video shows how a brainlife.io publication can be opened from the Data Deposition page of the journal Nature Scientific Data.
Difficulty level: Beginner
Duration: 2:25
Speaker: :
Course:
An introduction to data management, manipulation, visualization, and analysis for neuroscience. Students will learn scientific programming in Python, and use this to work with example data from areas such as cognitive-behavioral research, single-cell recording, EEG, and structural and functional MRI. Basic signal processing techniques including filtering are covered. The course includes a Jupyter Notebook and video tutorials.
Difficulty level: Beginner
Duration: 1:09:16
Speaker: : Aaron J. Newman
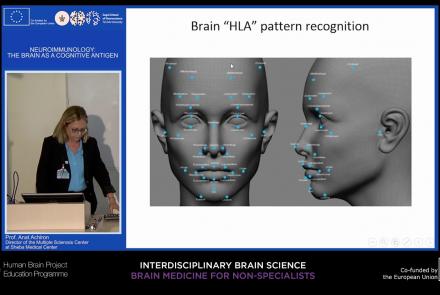
This lecture focuses on how the immune system can target and attack the nervous system to produce autoimmune responses that may result in diseases such as multiple sclerosis, neuromyelitis, and lupus cerebritis manifested by motor, sensory, and cognitive impairments. Despite the fact that the brain is an immune-privileged site, autoreactive lymphocytes producing proinflammatory cytokines can cause active brain inflammation, leading to myelin and axonal loss.
Difficulty level: Beginner
Duration: 37:36
Speaker: : Anat Achiron
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- (-) Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- (-) Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)