Lesson type
Difficulty level
This lecture describes how to build research workflows, including a demonstrate using DataJoint Elements to build data pipelines.
Difficulty level: Intermediate
Duration: 47:00
Speaker: : Dimitri Yatsenko
This lesson provides an introduction to the Symposium on Science Management at the Canadian Association for Neuroscience 2019 Meeting.
Difficulty level: Beginner
Duration: 9:52
Speaker: : Randy McIntosh
This lesson gives a primer to project management in a scientific context, with a particular neuroinformatic case study.
Difficulty level: Beginner
Duration: 19:06
Speaker: : Kelly Shen
In this lesson, you will hear about the current challenges regarding data management, as well as policies and resources aimed to address them.
Difficulty level: Beginner
Duration: 18:13
Speaker: : Mojib Javadi
This lesson covers "Knowledge Translation", the activities involved in moving research from the laboratory, the research journal, and the academic conference into the hands of people and organizations who can put it to practical use.
Difficulty level: Beginner
Duration: 15:05
Speaker: : Jordan Antflick
In this lesson, you will hear about the various methods developed and employed in managing performance.
Difficulty level: Beginner
Duration: 12:57
Speaker: : Christa Studzinski
This lesson provides an overview of how to manage relationships in a research context, while highlighting the need for effective communication at various levels.
Difficulty level: Beginner
Duration:
Speaker: : Helena Ledmyr
In this lesson you will hear a panel discussion which hosts experts in the field whom have extensive experience with management in a science setting.
Difficulty level: Beginner
Duration: 54:38
Speaker: :
This lesson provides an overview of the current status in the field of neuroscientific ontologies, presenting examples of data organization and standards, particularly from neuroimaging and electrophysiology.
Difficulty level: Intermediate
Duration: 33:41
Speaker: : Yaroslav O. Halchenko
This lesson continues from part one of the lecture Ontologies, Databases, and Standards, diving deeper into a description of ontologies and knowledg graphs.
Difficulty level: Intermediate
Duration: 50:18
Speaker: : Jeff Grethe
This lecture covers the NIDM data format within BIDS to make your datasets more searchable, and how to optimize your dataset searches.
Difficulty level: Beginner
Duration: 12:33
Speaker: : David Keator
This lecture covers positron emission tomography (PET) imaging and the Brain Imaging Data Structure (BIDS), and how they work together within the PET-BIDS standard to make neuroscience more open and FAIR.
Difficulty level: Beginner
Duration: 12:06
Speaker: : Melanie Ganz
This lecture discusses the FAIR principles as they apply to electrophysiology data and metadata, the building blocks for community tools and standards, platforms and grassroots initiatives, and the challenges therein.
Difficulty level: Beginner
Duration: 8:11
Speaker: : Thomas Wachtler
This lecture discusses how to standardize electrophysiology data organization to move towards being more FAIR.
Difficulty level: Beginner
Duration: 15:51
Speaker: : Sylvain Takerkart
Course:
The International Brain Initiative (IBI) is a consortium of the world’s major large-scale brain initiatives and other organizations with a vested interest in catalyzing and advancing neuroscience research through international collaboration and knowledge sharing. This workshop introduces the IBI, the efforts of the Data Standards and Sharing Working Group, and keynote lectures on the impact of data standards and sharing on large-scale brain projects, as well as a discussion on prospects and needs for neural data sharing.
Difficulty level: Intermediate
Duration: 2:06:58
The lesson introduces the Brain Imaging Data Structure (BIDS), the community standard for organizing, curating, and sharing neuroimaging and associated data. The session focuses on understanding the BIDS framework, learning its data structure and validation processes.
Difficulty level: Intermediate
Duration: 38:52
Speaker: : Cyril Pernet
This session moves from BIDS basics into analysis workflows, focusing on how to turn raw, BIDS-organized data into derivatives using BIDS Apps and containers for reproducible processing. It compares end-to-end pipelines across fMRI and PET (and notes EEG/MEG), explains typical preprocessing choices, and shows how standardized inputs plus containerized tools (Docker/AppTainer) yield consistent, auditable outputs.
Difficulty level: Intermediate
Duration: 56:03
Speaker: : Martin Nørgaard
The session explains GDPR rules around data sharing for research in Europe, the distinction between law and ethics, and introduces practical solutions for securely sharing sensitive datasets. Researchers have more flexibility than commonly assumed: scientific research is considered a public interest task, so explicit consent for data sharing isn’t legally required, though transparency and informing participants remain ethically important. The talk also introduces publicneuro.eu, a controlled-access platform that enables sharing neuroimaging datasets with open metadata, DOIs, and customizable access restrictions while ensuring GDPR compliance.
Difficulty level: Intermediate
Duration: 31:12
Speaker: : Cyril Pernet
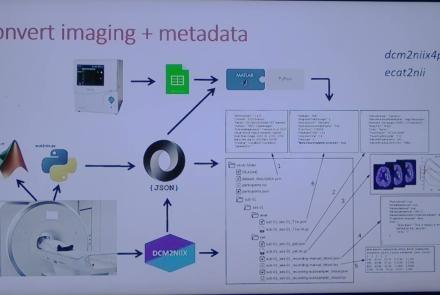
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 41:04
Speaker: : Martin Nørgaard
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- (-) Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)