Difficulty level
This lecture provides an introduction to the course "Cognitive Science & Psychology: Mind, Brain, and Behavior".
Difficulty level: Beginner
Duration: 1:06:49
Speaker: : Paul F.M.J. Verschure
This lesson covers the history of neuroscience and machine learning, and the story of how these two seemingly disparate fields are increasingly merging.
Difficulty level: Beginner
Duration: 12:25
Speaker: : Dan Goodman
In this lesson you will learn how machine learners and neuroscientists construct abstract computational models based on various neurophysiological signalling properties.
Difficulty level: Intermediate
Duration: 10:52
Speaker: : Dan Goodman
In this lesson, you will learn about some typical neuronal models employed by machine learners and computational neuroscientists, meant to imitate the biophysical properties of real neurons.
Difficulty level: Intermediate
Duration: 3:12
Speaker: : Dan Goodman
Whereas the previous two lessons described the biophysical and signalling properties of individual neurons, this lesson describes properties of those units when part of larger networks.
Difficulty level: Intermediate
Duration: 6:00
Speaker: : Marcus Ghosh
This lesson goes over some examples of how machine learners and computational neuroscientists go about designing and building neural network models inspired by biological brain systems.
Difficulty level: Intermediate
Duration: 12:52
Speaker: : Dan Goodman
In this lesson, you will learn about different approaches to modeling learning in neural networks, particularly focusing on system parameters such as firing rates and synaptic weights impact a network.
Difficulty level: Intermediate
Duration: 9:40
Speaker: : Dan Goodman
In this lesson, you will learn more about some of the issues inherent in modeling neural spikes, approaches to ameliorate these problems, and the pros and cons of these approaches.
Difficulty level: Intermediate
Duration: 5:31
Speaker: : Dan Goodman
In this lesson, you will learn about some of the many methods to train spiking neural networks (SNNs) with either no attempt to use gradients, or only use gradients in a limited or constrained way.
Difficulty level: Intermediate
Duration: 5:14
Speaker: : Dan Goodman
In this lesson, you will learn how to train spiking neural networks (SNNs) with a surrogate gradient method.
Difficulty level: Intermediate
Duration: 11:23
Speaker: : Dan Goodman
This lesson explores how researchers try to understand neural networks, particularly in the case of observing neural activity.
Difficulty level: Intermediate
Duration: 8:20
Speaker: : Marcus Ghosh
In this lesson you will learn about the motivation behind manipulating neural activity, and what forms that may take in various experimental designs.
Difficulty level: Intermediate
Duration: 8:42
Speaker: : Marcus Ghosh
This video briefly goes over the exercises accompanying Week 6 of the Neuroscience for Machine Learners (Neuro4ML) course, Understanding Neural Networks.
Difficulty level: Intermediate
Duration: 2:43
Speaker: : Marcus Ghosh
This lecture focuses on the structured validation process within computational neuroscience, including the tools, services, and methods involved in simulation and analysis.
Difficulty level: Beginner
Duration: 14:19
Speaker: : Michael Denker
Course:
This module explains how neurons come together to create the networks that give rise to our thoughts. The totality of our neurons and their connection is called our connectome. Learn how this connectome changes as we learn, and computes information.
Difficulty level: Beginner
Duration: 7:13
Speaker: : Harrison Canning
This lesson provides an overview of the current status in the field of neuroscientific ontologies, presenting examples of data organization and standards, particularly from neuroimaging and electrophysiology.
Difficulty level: Intermediate
Duration: 33:41
Speaker: : Yaroslav O. Halchenko
The lesson introduces the Brain Imaging Data Structure (BIDS), the community standard for organizing, curating, and sharing neuroimaging and associated data. The session focuses on understanding the BIDS framework, learning its data structure and validation processes.
Difficulty level: Intermediate
Duration: 38:52
Speaker: : Cyril Pernet
This session moves from BIDS basics into analysis workflows, focusing on how to turn raw, BIDS-organized data into derivatives using BIDS Apps and containers for reproducible processing. It compares end-to-end pipelines across fMRI and PET (and notes EEG/MEG), explains typical preprocessing choices, and shows how standardized inputs plus containerized tools (Docker/AppTainer) yield consistent, auditable outputs.
Difficulty level: Intermediate
Duration: 56:03
Speaker: : Martin Nørgaard
The session explains GDPR rules around data sharing for research in Europe, the distinction between law and ethics, and introduces practical solutions for securely sharing sensitive datasets. Researchers have more flexibility than commonly assumed: scientific research is considered a public interest task, so explicit consent for data sharing isn’t legally required, though transparency and informing participants remain ethically important. The talk also introduces publicneuro.eu, a controlled-access platform that enables sharing neuroimaging datasets with open metadata, DOIs, and customizable access restrictions while ensuring GDPR compliance.
Difficulty level: Intermediate
Duration: 31:12
Speaker: : Cyril Pernet
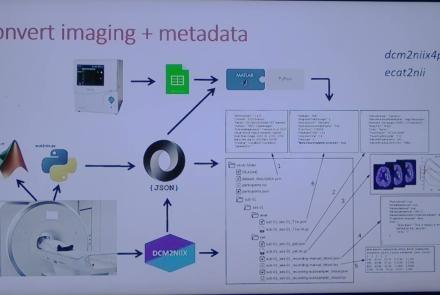
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- (-) Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)