Lesson type
Difficulty level
This video gives a brief introduction to Neuro4ML's lessons on neuromorphic computing - the use of specialized hardware which either directly mimics brain function or is inspired by some aspect of the way the brain computes.
Difficulty level: Intermediate
Duration: 3:56
Speaker: : Dan Goodman
In this lesson, you will learn in more detail about neuromorphic computing, that is, non-standard computational architectures that mimic some aspect of the way the brain works.
Difficulty level: Intermediate
Duration: 10:08
Speaker: : Dan Goodman
This video provides a very quick introduction to some of the neuromorphic sensing devices, and how they offer unique, low-power applications.
Difficulty level: Intermediate
Duration: 2:37
Speaker: : Dan Goodman
Course:
This lecture covers modeling the neuron in silicon, modeling vision and audition, and sensory fusion using a deep network.
Difficulty level: Beginner
Duration: 1:32:17
Speaker: : Shih-Chii Liu
This lesson presents a simulation software for spatial model neurons and their networks designed primarily for GPUs.
Difficulty level: Intermediate
Duration: 21:15
Speaker: : Tadashi Yamazaki
This lesson gives an overview of past and present neurocomputing approaches and hybrid analog/digital circuits that directly emulate the properties of neurons and synapses.
Difficulty level: Beginner
Duration: 41:57
Speaker: : Giacomo Indiveri
Presentation of the Brian neural simulator, where models are defined directly by their mathematical equations and code is automatically generated for each specific target.
Difficulty level: Beginner
Duration: 20:39
Speaker: : Giacomo Indiveri
The lecture covers a brief introduction to neuromorphic engineering, some of the neuromorphic networks that the speaker has developed, and their potential applications, particularly in machine learning.
Difficulty level: Intermediate
Duration: 19:57
Speaker: : Runchun Mark Wang
This lesson explains the fundamental principles of neuronal communication, such as neuronal spiking, membrane potentials, and cellular excitability, and how these electrophysiological features of the brain may be modelled and simulated digitally.
Difficulty level: Intermediate
Duration: 1:20:42
Speaker: : Etay Hay
This is a tutorial on how to simulate neuronal spiking in brain microcircuit models, as well as how to analyze, plot, and visualize the corresponding data.
Difficulty level: Intermediate
Duration: 1:39:50
Speaker: : Frank Mazza
This is an in-depth guide on EEG signals and their interaction within brain microcircuits. Participants are also shown techniques and software for simulating, analyzing, and visualizing these signals.
Difficulty level: Intermediate
Duration: 1:30:41
Speaker: : Frank Mazza
Course:
In this tutorial on simulating whole-brain activity using Python, participants can follow along using corresponding code and repositories, learning the basics of neural oscillatory dynamics, evoked responses and EEG signals, ultimately leading to the design of a network model of whole-brain anatomical connectivity.
Difficulty level: Intermediate
Duration: 1:16:10
Speaker: : John Griffiths
This tutorial walks participants through the application of dynamic causal modelling (DCM) to fMRI data using MATLAB. Participants are also shown various forms of DCM, how to generate and specify different models, and how to fit them to simulated neural and BOLD data.
This lesson corresponds to slides 158-187 of the PDF below.
Difficulty level: Advanced
Duration: 1:22:10
Speaker: : Peter Bedford, Povilas Karvelis
This lecture focuses on the structured validation process within computational neuroscience, including the tools, services, and methods involved in simulation and analysis.
Difficulty level: Beginner
Duration: 14:19
Speaker: : Michael Denker
Course:
This session will include presentations of infrastructure that embrace the FAIR principles developed by members of the INCF Community.
This lecture provides an overview of The Virtual Brain Simulation Platform.
Difficulty level: Beginner
Duration: 9:36
Speaker: : Petra Ritter
Course:
This tutorial demonstrates how to use PyNN, a simulator-independent language for building neuronal network models, in conjunction with the neuromorphic hardware system SpiNNaker.
Difficulty level: Intermediate
Duration: 25:49
Speaker: : Christian Brenninkmeijer
The lesson introduces the Brain Imaging Data Structure (BIDS), the community standard for organizing, curating, and sharing neuroimaging and associated data. The session focuses on understanding the BIDS framework, learning its data structure and validation processes.
Difficulty level: Intermediate
Duration: 38:52
Speaker: : Cyril Pernet
This session moves from BIDS basics into analysis workflows, focusing on how to turn raw, BIDS-organized data into derivatives using BIDS Apps and containers for reproducible processing. It compares end-to-end pipelines across fMRI and PET (and notes EEG/MEG), explains typical preprocessing choices, and shows how standardized inputs plus containerized tools (Docker/AppTainer) yield consistent, auditable outputs.
Difficulty level: Intermediate
Duration: 56:03
Speaker: : Martin Nørgaard
The session explains GDPR rules around data sharing for research in Europe, the distinction between law and ethics, and introduces practical solutions for securely sharing sensitive datasets. Researchers have more flexibility than commonly assumed: scientific research is considered a public interest task, so explicit consent for data sharing isn’t legally required, though transparency and informing participants remain ethically important. The talk also introduces publicneuro.eu, a controlled-access platform that enables sharing neuroimaging datasets with open metadata, DOIs, and customizable access restrictions while ensuring GDPR compliance.
Difficulty level: Intermediate
Duration: 31:12
Speaker: : Cyril Pernet
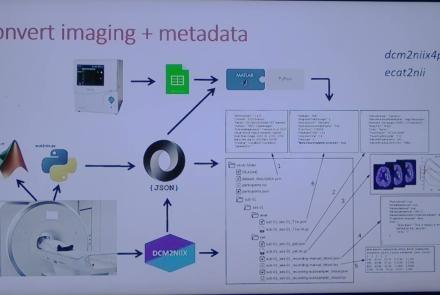
This session introduces the PET-to-BIDS (PET2BIDS) library, a toolkit designed to simplify the conversion and preparation of PET imaging datasets into BIDS-compliant formats. It supports multiple data types and formats (e.g., DICOM, ECAT7+, nifti, JSON), integrates seamlessly with Excel-based metadata, and provides automated routines for metadata updates, blood data conversion, and JSON synchronization. PET2BIDS improves human readability by mapping complex reconstruction names into standardized, descriptive labels and offers extensive documentation, examples, and video tutorials to make adoption easier for researchers.
Difficulty level: Intermediate
Duration: 9:23
Speaker: : Cyril Pernet
Topics
- Artificial Intelligence (7)
- Philosophy of Science (5)
- Provenance (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (8)
- Assembly 2021 (29)
- Brain-hardware interfaces (14)
- Clinical neuroscience (40)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(62)
- Neuroscience (11)
- Cognitive Science (7)
- Cell signaling (6)
- Brain networks (11)
- Glia (1)
- Electrophysiology (41)
- Learning and memory (5)
- Neuroanatomy (24)
- Neurobiology (16)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (15)
- Neurophysiology (27)
- Neuropharmacology (2)
- Neuronal plasticity (16)
- Synaptic plasticity (4)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(27)
- Computational neuroscience (279)
- Statistics (7)
- Computer Science (21)
- Genomics (34)
- Data science
(34)
- Open science (61)
- Project management (8)
- Education (4)
- Publishing (4)
- Neuroethics (42)