Lesson type
Difficulty level
This lecture provides an introduction to the course "Cognitive Science & Psychology: Mind, Brain, and Behavior".
Difficulty level: Beginner
Duration: 1:06:49
Speaker: : Paul F.M.J. Verschure
In this lesson, you will learn about different approaches to modeling learning in neural networks, particularly focusing on system parameters such as firing rates and synaptic weights impact a network.
Difficulty level: Intermediate
Duration: 9:40
Speaker: : Dan Goodman
In this lesson, you will learn about some of the many methods to train spiking neural networks (SNNs) with either no attempt to use gradients, or only use gradients in a limited or constrained way.
Difficulty level: Intermediate
Duration: 5:14
Speaker: : Dan Goodman
In this lesson, you will learn how to train spiking neural networks (SNNs) with a surrogate gradient method.
Difficulty level: Intermediate
Duration: 11:23
Speaker: : Dan Goodman
This lesson explores how researchers try to understand neural networks, particularly in the case of observing neural activity.
Difficulty level: Intermediate
Duration: 8:20
Speaker: : Marcus Ghosh
In this lesson you will learn about the motivation behind manipulating neural activity, and what forms that may take in various experimental designs.
Difficulty level: Intermediate
Duration: 8:42
Speaker: : Marcus Ghosh
This lecture focuses on the structured validation process within computational neuroscience, including the tools, services, and methods involved in simulation and analysis.
Difficulty level: Beginner
Duration: 14:19
Speaker: : Michael Denker
Course:
This module explains how neurons come together to create the networks that give rise to our thoughts. The totality of our neurons and their connection is called our connectome. Learn how this connectome changes as we learn, and computes information.
Difficulty level: Beginner
Duration: 7:13
Speaker: : Harrison Canning
This lesson discusses a gripping neuroscientific question: why have neurons developed the discrete action potential, or spike, as a principle method of communication?
Difficulty level: Intermediate
Duration: 9:34
Speaker: : Dan Goodman
This lesson provides an overview of Neurodata Without Borders (NWB), an ecosystem for neurophysiology data standardization. The lecture also introduces some NWB-enabled tools.
Difficulty level: Beginner
Duration: 29:53
Speaker: : Oliver Ruebel
Learn how to create a standard extracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 23:10
Speaker: : Ryan Ly
Learn how to create a standard calcium imaging dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 31:04
Speaker: : Ryan Ly
In this tutorial, you will learn how to create a standard intracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 20:23
Speaker: : Pamela Baker
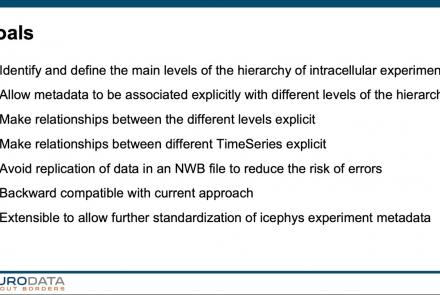
In this tutorial, you will learn how to use the icephys-metadata extension to enter meta-data detailing your experimental paradigm.
Difficulty level: Intermediate
Duration: 27:18
Speaker: : Oliver Ruebel
This lesson provides instructions on how to build and share extensions in NWB.
Difficulty level: Advanced
Duration: 20:29
Speaker: : Ryan Ly
Learn how to build custom APIs for extension.
Difficulty level: Advanced
Duration: 25:40
Speaker: : Andrew Tritt
This lesson provides instruction on advanced writing strategies in HDF5 that are accessible through PyNWB.
Difficulty level: Advanced
Duration: 23:00
Speaker: : Oliver Ruebel
In this tutorial, users learn how to create a standard extracellular electrophysiology dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 45:46
Speaker: : Ben Dichter
Learn how to create a standard calcium imaging dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 39:10
Speaker: : Ben Dichter
Learn how to create a standard intracellular electrophysiology dataset in NWB.
Difficulty level: Intermediate
Duration: 20:22
Speaker: : Pamela Baker
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Notebooks (1)
- Provenance (1)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (2)
- Assembly 2021 (27)
- Brain-hardware interfaces (13)
- Clinical neuroscience (23)
- International Brain Initiative (2)
- Repositories and science gateways (5)
- Resources (5)
- General neuroscience
(17)
- General neuroinformatics
(5)
- Computational neuroscience (101)
- Statistics (2)
- Computer Science (7)
- Genomics (3)
- Data science
(14)
- Open science (12)
- Project management (7)
- Education (1)
- Neuroethics (28)