Lesson type
Difficulty level
This lecture gives an introduction to the European Academy of Neurology, its recent achievements and ambitions.
Difficulty level: Intermediate
Duration: 21:57
Speaker: : Paul Boon
This lecture gives an overview on the European Health Dataspace.
Difficulty level: Intermediate
Duration: 26:33
Speaker: : Licino Kustra Mano
This lecture covers a wide range of aspects regarding neuroinformatics and data governance, describing both their historical developments and current trajectories. Particular tools, platforms, and standards to make your research more FAIR are also discussed.
Difficulty level: Beginner
Duration: 54:58
Speaker: : Franco Pestilli
Course:
This lecture covers the needs and challenges involved in creating a FAIR ecosystem for neuroimaging research.
Difficulty level: Beginner
Duration: 12:26
Speaker: : Camille Maumet
Course:
This lecture covers multiple aspects of FAIR neuroscience data: what makes it unique, the challenges to making it FAIR, the importance of overcoming these challenges, and how data governance comes into play.
Difficulty level: Beginner
Duration: 14:56
Speaker: : Damian Eke
This lecture covers how you can make your data public through EBRAINS. This talk focuses on the ethical considerations for sharing data, the requirements that are imposed by various regulations, particularly for sharing human data. The lecture also includes a discussion of how EBRAINS designs its services to deal with the ethical and regulatory aspects of sharing these kinds of data.
Difficulty level: Intermediate
Duration: 16:15
Speaker: : Maaike van Swieten and Jan Bjaalie
This lecture discusses differential privacy and synthetic data in the context of medical data sharing in clinical neurosciences.
Difficulty level: Intermediate
Duration: 20:26
Speaker: : Minos Garofalakis
Learn how to create a standard extracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 23:10
Speaker: : Ryan Ly
Learn how to create a standard calcium imaging dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 31:04
Speaker: : Ryan Ly
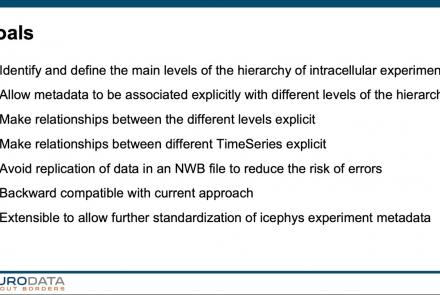
In this tutorial, you will learn how to create a standard intracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 20:23
Speaker: : Pamela Baker
In this tutorial, you will learn how to use the icephys-metadata extension to enter meta-data detailing your experimental paradigm.
Difficulty level: Intermediate
Duration: 27:18
Speaker: : Oliver Ruebel
This lesson provides instructions on how to build and share extensions in NWB.
Difficulty level: Advanced
Duration: 20:29
Speaker: : Ryan Ly
Learn how to build custom APIs for extension.
Difficulty level: Advanced
Duration: 25:40
Speaker: : Andrew Tritt
This lesson provides instruction on advanced writing strategies in HDF5 that are accessible through PyNWB.
Difficulty level: Advanced
Duration: 23:00
Speaker: : Oliver Ruebel
In this tutorial, users learn how to create a standard extracellular electrophysiology dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 45:46
Speaker: : Ben Dichter
Learn how to create a standard calcium imaging dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 39:10
Speaker: : Ben Dichter
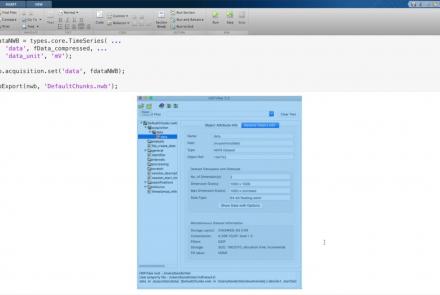
Learn how to create a standard intracellular electrophysiology dataset in NWB.
Difficulty level: Intermediate
Duration: 20:22
Speaker: : Pamela Baker
This lesson provides a tutorial on how to handle writing very large data in MatNWB.
Difficulty level: Advanced
Duration: 16:18
Speaker: : Ben Dichter
This lesson gives an overview of the Brainstorm package for analyzing extracellular electrophysiology, including preprocessing, spike sorting, trial alignment, and spectrotemporal decomposition.
Difficulty level: Intermediate
Duration: 47:47
Speaker: : Konstantinos Nasiotis
This lesson provides an overview of the CaImAn package, as well as a demonstration of usage with NWB.
Difficulty level: Intermediate
Duration: 44:37
Speaker: : Andrea Giovannucci
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Notebooks (1)
- Provenance (1)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (2)
- Assembly 2021 (27)
- Brain-hardware interfaces (13)
- Clinical neuroscience (23)
- International Brain Initiative (2)
- Repositories and science gateways (5)
- Resources (5)
- General neuroscience
(17)
- General neuroinformatics
(5)
- Computational neuroscience (101)
- Statistics (2)
- Computer Science (7)
- Genomics (3)
- Data science
(14)
- Open science (12)
- Project management (7)
- Education (1)
- Neuroethics (28)