Lesson type
Difficulty level
Course:
This lesson introduces the EEGLAB toolbox, as well as motivations for its use.
Difficulty level: Beginner
Duration: 15:32
Speaker: : Arnaud Delorme
Course:
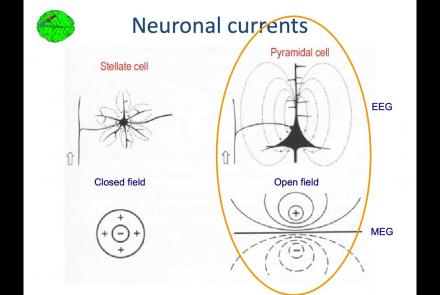
In this lesson, you will learn about the biological activity which generates and is measured by the EEG signal.
Difficulty level: Beginner
Duration: 6:53
Speaker: : Arnaud Delorme
Course:
This lesson goes over the characteristics of EEG signals when analyzed in source space (as opposed to sensor space).
Difficulty level: Beginner
Duration: 10:56
Speaker: : Arnaud Delorme
Course:
This lesson describes the development of EEGLAB as well as to what extent it is used by the research community.
Difficulty level: Beginner
Duration: 6:06
Speaker: : Arnaud Delorme
Course:
This lesson provides instruction as to how to build a processing pipeline in EEGLAB for a single participant.
Difficulty level: Beginner
Duration: 9:20
Speaker: :
Course:
Whereas the previous lesson of this course outlined how to build a processing pipeline for a single participant, this lesson discusses analysis pipelines for multiple participants simultaneously.
Difficulty level: Beginner
Duration: 10:55
Speaker: : Arnaud Delorme
Course:
In addition to outlining the motivations behind preprocessing EEG data in general, this lesson covers the first step in preprocessing data with EEGLAB, importing raw data.
Difficulty level: Beginner
Duration: 8:30
Speaker: : Arnaud Delorme
Course:
Continuing along the EEGLAB preprocessing pipeline, this tutorial walks users through how to import data events as well as EEG channel locations.
Difficulty level: Beginner
Duration: 11:53
Speaker: : Arnaud Delorme
Course:
This tutorial demonstrates how to re-reference and resample raw data in EEGLAB, why such steps are important or useful in the preprocessing pipeline, and how choices made at this step may affect subsequent analyses.
Difficulty level: Beginner
Duration: 11:48
Speaker: : Arnaud Delorme
Course:
This tutorial instructs users how to visually inspect partially pre-processed neuroimaging data in EEGLAB, specifically how to use the data browser to investigate specific channels, epochs, or events for removable artifacts, biological (e.g., eye blinks, muscle movements, heartbeat) or otherwise (e.g., corrupt channel, line noise).
Difficulty level: Beginner
Duration: 5:08
Speaker: : Arnaud Delorme
Course:
This tutorial provides instruction on how to use EEGLAB to further preprocess EEG datasets by identifying and discarding bad channels which, if left unaddressed, can corrupt and confound subsequent analysis steps.
Difficulty level: Beginner
Duration: 13:01
Speaker: : Arnaud Delorme
Course:
Users following this tutorial will learn how to identify and discard bad EEG data segments using the MATLAB toolbox EEGLAB.
Difficulty level: Beginner
Duration: 11:25
Speaker: : Arnaud Delorme
Course:
An introduction to data management, manipulation, visualization, and analysis for neuroscience. Students will learn scientific programming in Python, and use this to work with example data from areas such as cognitive-behavioral research, single-cell recording, EEG, and structural and functional MRI. Basic signal processing techniques including filtering are covered. The course includes a Jupyter Notebook and video tutorials.
Difficulty level: Beginner
Duration: 1:09:16
Speaker: : Aaron J. Newman
This tutorial demonstrates how to perform cell-type deconvolution in order to estimate how proportions of cell-types in the brain change in response to various conditions. While these techniques may be useful in addressing a wide range of scientific questions, this tutorial will focus on the cellular changes associated with major depression (MDD).
Difficulty level: Intermediate
Duration: 1:15:14
Speaker: : Keon Arbabi
Course:
This video shows how to use the brainlife.io interface to edit the participants' info file. This file is the ParticipantInfo.json file of the Brain Imaging Data Structure (BIDS).
Difficulty level: Beginner
Duration: 0:34
Speaker: :
Course:
This video will document the process of running an app on brainlife, from data staging to archiving of the final data outputs.
Difficulty level: Beginner
Duration: 3:43
Speaker: :
Course:
This video demonstrates each required step for preprocessing T1w anatomical data in brainlife.io.
Difficulty level: Beginner
Duration: 3:28
Speaker: :
Course:
This short video shows how data in a brainlife.io publication can be opened from a DOI inside a published article. The video provides an example of how the DOI deposited on the journal can be opened with a web browser to redirect to the associated data publication on brainlife.io.
Difficulty level: Beginner
Duration: 2:18
Speaker: :
This video explains what metadata is, why it is important, and how you can organize your metadata to increase the FAIRness of your data on EBRAINS.
Difficulty level: Beginner
Duration: 17:23
Speaker: : Ulrike Schlegel
Course:
This lecture covers the rationale for developing the DAQCORD, a framework for the design, documentation, and reporting of data curation methods in order to advance the scientific rigour, reproducibility, and analysis of data.
Difficulty level: Intermediate
Duration: 17:08
Speaker: : Ari Ercole
Topics
- Standards and Best Practices (2)
- Notebooks (2)
- Clinical neuroinformatics (2)
- Provenance (2)
- Artificial Intelligence (1)
- Digital brain atlasing (3)
- Neuroimaging (36)
- Optogenetics (1)
- Standards and best practices (1)
- Tools (20)
- Workflows (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (1)
- Assembly 2021 (1)
- Brain-hardware interfaces (12)
- Clinical neuroscience (1)
- Repositories and science gateways (1)
- Resources (1)
- General neuroscience (11)
- Phenome (1)
- Computational neuroscience (80)
- Statistics (5)
- Computer Science (4)
- Genomics (28)
- Data science (13)
- Open science (18)
- Education (1)
- Neuroethics (1)