Lesson type
Difficulty level
Course:
This lecture covers the description and characterization of an input-output relationship in a information-theoretic context.
Difficulty level: Beginner
Duration: 1:35:33
Speaker: : Jonathan D. Victor
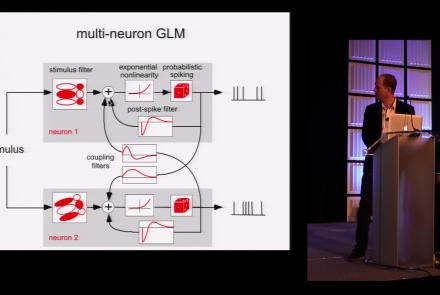
This lesson is part 1 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:45:48
Speaker: : Jonathan Pillow
This lesson is part 2 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:50:31
Speaker: : Jonathan Pillow
Course:
From the retina to the superior colliculus, the lateral geniculate nucleus into primary visual cortex and beyond, this lecture gives a tour of the mammalian visual system highlighting the Nobel-prize winning discoveries of Hubel & Wiesel.
Difficulty level: Beginner
Duration: 56:31
Speaker: : Clay Reid
Course:
From Universal Turing Machines to McCulloch-Pitts and Hopfield associative memory networks, this lecture explains what is meant by computation.
Difficulty level: Beginner
Duration: 55:27
Speaker: : Christof Koch
In this lesson you will learn about ion channels and the movement of ions across the cell membrane, one of the key mechanisms underlying neuronal communication.
Difficulty level: Beginner
Duration: 25:51
Speaker: : Carl Petersen
This lecture provides an introduction to the application of genetic testing in neurodevelopmental disorders.
Difficulty level: Beginner
Duration: 37:47
Speaker: : Diana-Laura Miclea
In this lesson, you will learn about how genetics can contribute to our understanding of psychiatric phenotypes.
Difficulty level: Beginner
Duration: 55:15
Speaker: : Sven Cichon
The Allen Mouse Brain Atlas is a genome-wide, high-resolution atlas of gene expression throughout the adult mouse brain. This tutorial describes the basic search and navigation features of the Allen Mouse Brain Atlas.
Difficulty level: Beginner
Duration: 6:40
Speaker: : Allen Institute for Brain Science
The Allen Developing Mouse Brain Atlas is a detailed atlas of gene expression across mouse brain development. This tutorial describes the basic search and navigation features of the Allen Developing Mouse Brain Atlas.
Difficulty level: Beginner
Duration: 6:35
Speaker: : Unknown
This tutorial demonstrates how to use the differential search feature of the Allen Mouse Brain Atlas to find gene markers for different regions of the brain, as well as to visualize this gene expression in three-dimensional space. Differential search is also available for the Allen Developing Mouse Brain Atlas and the Allen Human Brain Atlas.
Difficulty level: Beginner
Duration: 6:31
Speaker: : Unknown
Course:
This lesson provides an overview of GeneWeaver, a web application for the integrated cross-species analysis of functional genomics data to find convergent evidence from heterogeneous sources.
Difficulty level: Beginner
Duration: 1:03:26
Speaker: : Erich J. Baker
Course:
This lesson provides a demonstration of GeneWeaver, a system for the integration and analysis of heterogeneous functional genomics data.
Difficulty level: Beginner
Duration: 25:53
Speaker: :
Course:
This lecture outlines GeneNetwork.org, a group of linked data sets and tools used to study complex networks of genes, molecules, and higher order gene function and phenotypes.
Difficulty level: Beginner
Duration: 1:00:43
Speaker: : Robert Williams
Course:
This tutorial shows how to use the UCSC genome browser to find a list of genes in a given genomic region.
Difficulty level: Beginner
Duration: 4:32
Speaker: : UCSC Genome Browser
Course:
This tutorial shows how to find all the single nucleotide polymorphisms (SNPs) upstream from genes using the UCSC Genome Browser.
Difficulty level: Beginner
Duration: 8:13
Speaker: : UCSC Genome Browser
Course:
This tutorial demonstrates how to find all the single nucleotide polymorphisms (SNPs) in a gene using the UCSC Genome Browser.
Difficulty level: Beginner
Duration: 6:12
Speaker: : UCSC Genome Browser
Course:
The Saved Sessions feature of the Browser has been around for quite some time, but many of our users have not made full use of it. This feature offers a great way to keep track of your thinking on a particular topic.
Difficulty level: Beginner
Duration: 7:16
Speaker: : UCSC Genome Browser
Course:
The Track Collection Builder is a new tool in the UCSC Genome Browser that provides a way to create grouped collections of sub-tracks with native tracks, custom tracks, or hub tracks of continuous value graphing data types.
Difficulty level: Beginner
Duration: 2:18
Speaker: : UCSC Genome Browser
Course:
This tutorial demonstrates the visibility controls on the Genome Browser, showing the effect on BED tracks, wiggle tracks, and Conservation tracks. It also discusses supertracks and composite tracks.
Difficulty level: Beginner
Duration: 14:30
Speaker: : UCSC Genome Browser
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Provenance (2)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (6)
- Assembly 2021 (29)
- Brain-hardware interfaces (13)
- Clinical neuroscience (17)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(45)
- Neuroscience (9)
- Cognitive Science (7)
- Cell signaling (3)
- Brain networks (4)
- Glia (1)
- Electrophysiology (16)
- Learning and memory (3)
- Neuroanatomy (17)
- Neurobiology (7)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (4)
- Neurophysiology (22)
- Neuropharmacology (2)
- Synaptic plasticity (2)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(15)
- Computational neuroscience (195)
- Statistics (2)
- Computer Science (15)
- (-) Genomics (26)
- Data science
(24)
- Open science (56)
- Project management (7)
- Education (3)
- Publishing (4)
- Neuroethics (37)