Lesson type
Difficulty level
Course:
This lecture covers the description and characterization of an input-output relationship in a information-theoretic context.
Difficulty level: Beginner
Duration: 1:35:33
Speaker: : Jonathan D. Victor
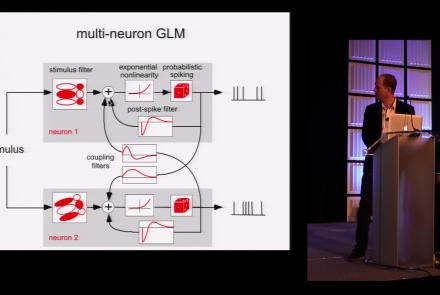
This lesson is part 1 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:45:48
Speaker: : Jonathan Pillow
This lesson is part 2 of 2 of a tutorial on statistical models for neural data.
Difficulty level: Beginner
Duration: 1:50:31
Speaker: : Jonathan Pillow
Course:
From the retina to the superior colliculus, the lateral geniculate nucleus into primary visual cortex and beyond, this lecture gives a tour of the mammalian visual system highlighting the Nobel-prize winning discoveries of Hubel & Wiesel.
Difficulty level: Beginner
Duration: 56:31
Speaker: : Clay Reid
Course:
From Universal Turing Machines to McCulloch-Pitts and Hopfield associative memory networks, this lecture explains what is meant by computation.
Difficulty level: Beginner
Duration: 55:27
Speaker: : Christof Koch
In this lesson you will learn about ion channels and the movement of ions across the cell membrane, one of the key mechanisms underlying neuronal communication.
Difficulty level: Beginner
Duration: 25:51
Speaker: : Carl Petersen
Course:
This video will document the process of running an app on brainlife, from data staging to archiving of the final data outputs.
Difficulty level: Beginner
Duration: 3:43
Speaker: :
Course:
This quick video presents some of the various visualizers available on brainlife.io
Difficulty level: Beginner
Duration: 1:11
Speaker: :
Course:
This short video shows how a brainlife.io publication can be opened from the Data Deposition page of the journal Nature Scientific Data.
Difficulty level: Beginner
Duration: 2:25
Speaker: :
Course:
An introduction to data management, manipulation, visualization, and analysis for neuroscience. Students will learn scientific programming in Python, and use this to work with example data from areas such as cognitive-behavioral research, single-cell recording, EEG, and structural and functional MRI. Basic signal processing techniques including filtering are covered. The course includes a Jupyter Notebook and video tutorials.
Difficulty level: Beginner
Duration: 1:09:16
Speaker: : Aaron J. Newman
Topics
- Artificial Intelligence (6)
- Philosophy of Science (5)
- Provenance (2)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (6)
- Assembly 2021 (29)
- Brain-hardware interfaces (13)
- Clinical neuroscience (17)
- International Brain Initiative (2)
- Repositories and science gateways (11)
- Resources (6)
- General neuroscience
(45)
- Neuroscience (9)
- Cognitive Science (7)
- Cell signaling (3)
- Brain networks (4)
- Glia (1)
- Electrophysiology (16)
- Learning and memory (3)
- Neuroanatomy (17)
- Neurobiology (7)
- Neurodegeneration (1)
- Neuroimmunology (1)
- Neural networks (4)
- Neurophysiology (22)
- Neuropharmacology (2)
- Synaptic plasticity (2)
- Visual system (12)
- Phenome (1)
- General neuroinformatics
(15)
- Computational neuroscience (195)
- Statistics (2)
- Computer Science (15)
- Genomics (26)
- Data science
(24)
- Open science (56)
- Project management (7)
- Education (3)
- Publishing (4)
- Neuroethics (37)