This lecture presents the Medical Informatics Platform's data federation in epilepsy.
Difficulty level: Intermediate
Duration: 27:09
Speaker: : Philippe Ryvlin
This lecture aims to help researchers, students, and health care professionals understand the place for neuroinformatics in the patient journey using the exemplar of an epilepsy patient.
Difficulty level: Intermediate
Duration: 1:32:53
Speaker: : Randy Gollub & Prantik Kundu
Explore how to setup an epileptic seizure simulation with the TVB graphical user interface. This lesson will show you how to program the epileptor model in the brain network to simulate a epileptic seizure originating in the hippocampus. It will also show how to upload and view mouse connectivity data, as well as give a short introduction to the python script interface of TVB.
Difficulty level: Intermediate
Duration: 58:06
Speaker: : Paul Triebkorn
This talk introduces data sharing initiatives in Epilepsy, particularly across Europe.
Difficulty level: Intermediate
Duration: 13:56
Speaker: : J. Helen Cross
This lecture provides an introduction to the Brain Imaging Data Structure (BIDS), a standard for organizing human neuroimaging datasets.
Difficulty level: Intermediate
Duration: 56:49
Speaker: : Chris Gorgolewski
Course:
This lesson outlines Neurodata Without Borders (NWB), a data standard for neurophysiology which provides neuroscientists with a common standard to share, archive, use, and build analysis tools for neurophysiology data.
Difficulty level: Intermediate
Duration: 29:53
Speaker: : Oliver Ruebel
Course:
This lecture covers the rationale for developing the DAQCORD, a framework for the design, documentation, and reporting of data curation methods in order to advance the scientific rigour, reproducibility, and analysis of data.
Difficulty level: Intermediate
Duration: 17:08
Speaker: : Ari Ercole
Course:
This tutorial demonstrates how to use PyNN, a simulator-independent language for building neuronal network models, in conjunction with the neuromorphic hardware system SpiNNaker.
Difficulty level: Intermediate
Duration: 25:49
Speaker: : Christian Brenninkmeijer
In this lesson, you will learn in more detail about neuromorphic computing, that is, non-standard computational architectures that mimic some aspect of the way the brain works.
Difficulty level: Intermediate
Duration: 10:08
Speaker: : Dan Goodman
This video provides a very quick introduction to some of the neuromorphic sensing devices, and how they offer unique, low-power applications.
Difficulty level: Intermediate
Duration: 2:37
Speaker: : Dan Goodman
This lecture gives an overview of how to prepare and preprocess neuroimaging (EEG/MEG) data for use in TVB.
Difficulty level: Intermediate
Duration: 1:40:52
Speaker: : Paul Triebkorn
Course:
This lesson is a general overview of overarching concepts in neuroinformatics research, with a particular focus on clinical approaches to defining, measuring, studying, diagnosing, and treating various brain disorders. Also described are the complex, multi-level nature of brain disorders and the data associated with them, from genes and individual cells up to cortical microcircuits and whole-brain network dynamics. Given the heterogeneity of brain disorders and their underlying mechanisms, this lesson lays out a case for multiscale neuroscience data integration.
Difficulty level: Intermediate
Duration: 1:09:33
Speaker: : Sean Hill
Course:
In this tutorial on simulating whole-brain activity using Python, participants can follow along using corresponding code and repositories, learning the basics of neural oscillatory dynamics, evoked responses and EEG signals, ultimately leading to the design of a network model of whole-brain anatomical connectivity.
Difficulty level: Intermediate
Duration: 1:16:10
Speaker: : John Griffiths
This lesson breaks down the principles of Bayesian inference and how it relates to cognitive processes and functions like learning and perception. It is then explained how cognitive models can be built using Bayesian statistics in order to investigate how our brains interface with their environment.
This lesson corresponds to slides 1-64 in the PDF below.
Difficulty level: Intermediate
Duration: 1:28:14
Speaker: : Andreea Diaconescu
Course:
This lecture and tutorial focuses on measuring human functional brain networks, as well as how to account for inherent variability within those networks.
Difficulty level: Intermediate
Duration: 50:44
Speaker: : Caterina Gratton
Learn how to create a standard extracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 23:10
Speaker: : Ryan Ly
Learn how to create a standard calcium imaging dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 31:04
Speaker: : Ryan Ly
In this tutorial, you will learn how to create a standard intracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 20:23
Speaker: : Pamela Baker
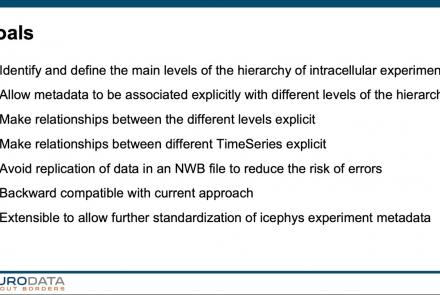
In this tutorial, you will learn how to use the icephys-metadata extension to enter meta-data detailing your experimental paradigm.
Difficulty level: Intermediate
Duration: 27:18
Speaker: : Oliver Ruebel
In this tutorial, users learn how to create a standard extracellular electrophysiology dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 45:46
Speaker: : Ben Dichter
Topics
- Artificial Intelligence (1)
- Provenance (1)
- EBRAINS RI (6)
- Animal models (1)
- Brain-hardware interfaces (1)
- Clinical neuroscience (20)
- (-) General neuroscience (14)
- General neuroinformatics (1)
- Computational neuroscience (50)
- Statistics (5)
- Computer Science (3)
- Genomics (8)
- Data science (9)
- Open science (4)
- Project management (1)
- Neuroethics (3)