Course:
This tutorial demonstrates how to work with neuronal data using MATLAB, including actional potentials and spike counts, orientation tuing curves in visual cortex, and spatial maps of firing rates.
Difficulty level: Intermediate
Duration: 5:17
Speaker: : Mike X. Cohen
Course:
This lesson instructs users on how to import electrophysiological neural data into MATLAB, as well as how to convert spikes to a data matrix.
Difficulty level: Intermediate
Duration: 11:37
Speaker: : Mike X. Cohen
Course:
In this lesson, users will learn about human brain signals as measured by electroencephalography (EEG), as well as associated neural signatures such as steady state visually evoked potentials (SSVEPs) and alpha oscillations.
Difficulty level: Intermediate
Duration: 8:51
Speaker: : Mike X. Cohen
This lecture presents the Medical Informatics Platform's data federation in epilepsy.
Difficulty level: Intermediate
Duration: 27:09
Speaker: : Philippe Ryvlin
This is a tutorial introducing participants to the basics of RNA-sequencing data and how to analyze its features using Seurat.
Difficulty level: Intermediate
Duration: 1:19:17
Speaker: : Sonny Chen
This is a continuation of the talk on the cellular mechanisms of neuronal communication, this time at the level of brain microcircuits and associated global signals like those measureable by electroencephalography (EEG). This lecture also discusses EEG biomarkers in mental health disorders, and how those cortical signatures may be simulated digitally.
Difficulty level: Intermediate
Duration: 1:11:04
Speaker: : Etay Hay
This lecture aims to help researchers, students, and health care professionals understand the place for neuroinformatics in the patient journey using the exemplar of an epilepsy patient.
Difficulty level: Intermediate
Duration: 1:32:53
Speaker: : Randy Gollub & Prantik Kundu
In this final lecture of the INCF Short Course: Introduction to Neuroinformatics, you will hear about new advances in the application of machine learning methods to clinical neuroscience data. In particular, this talk discusses the performance of SynthSeg, an image segmentation tool for automated analysis of highly heterogeneous brain MRI clinical scans.
Difficulty level: Intermediate
Duration: 1:32:01
Speaker: : Juan Eugenio Iglesias
This lesson characterizes different types of learning in a neuroscientific and cellular context, and various models employed by researchers to investigate the mechanisms involved.
Difficulty level: Intermediate
Duration: 3:54
Speaker: : Dan Goodman
In this lesson, you will learn about different approaches to modeling learning in neural networks, particularly focusing on system parameters such as firing rates and synaptic weights impact a network.
Difficulty level: Intermediate
Duration: 9:40
Speaker: : Dan Goodman
This lesson describes spike timing-dependent plasticity (STDP), a biological process that adjusts the strength of connections between neurons in the brain, and how one can implement or mimic this process in a computational model. You will also find links for practical exercises at the bottom of this page.
Difficulty level: Intermediate
Duration: 12:50
Speaker: : Dan Goodman
In this lesson, you will learn about some of the many methods to train spiking neural networks (SNNs) with either no attempt to use gradients, or only use gradients in a limited or constrained way.
Difficulty level: Intermediate
Duration: 5:14
Speaker: : Dan Goodman
In this lesson, you will learn how to train spiking neural networks (SNNs) with a surrogate gradient method.
Difficulty level: Intermediate
Duration: 11:23
Speaker: : Dan Goodman
Explore how to setup an epileptic seizure simulation with the TVB graphical user interface. This lesson will show you how to program the epileptor model in the brain network to simulate a epileptic seizure originating in the hippocampus. It will also show how to upload and view mouse connectivity data, as well as give a short introduction to the python script interface of TVB.
Difficulty level: Intermediate
Duration: 58:06
Speaker: : Paul Triebkorn
Course:
This lesson gives an introduction to the central concepts of machine learning, and how they can be applied in Python using the scikit-learn package.
Difficulty level: Intermediate
Duration: 2:22:28
Speaker: : Jake Vanderplas
Learn how to create a standard extracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 23:10
Speaker: : Ryan Ly
Learn how to create a standard calcium imaging dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 31:04
Speaker: : Ryan Ly
In this tutorial, you will learn how to create a standard intracellular electrophysiology dataset in NWB using Python.
Difficulty level: Intermediate
Duration: 20:23
Speaker: : Pamela Baker
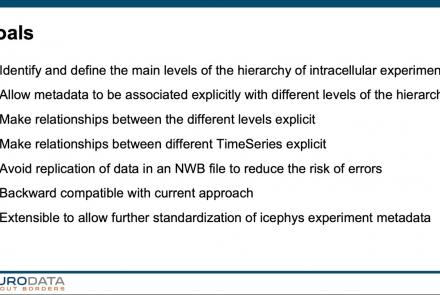
In this tutorial, you will learn how to use the icephys-metadata extension to enter meta-data detailing your experimental paradigm.
Difficulty level: Intermediate
Duration: 27:18
Speaker: : Oliver Ruebel
In this tutorial, users learn how to create a standard extracellular electrophysiology dataset in NWB using MATLAB.
Difficulty level: Intermediate
Duration: 45:46
Speaker: : Ben Dichter
Topics
- Artificial Intelligence (1)
- Provenance (1)
- EBRAINS RI (6)
- Brain-hardware interfaces (1)
- Clinical neuroscience (20)
- General neuroscience (9)
- General neuroinformatics (1)
- Computational neuroscience (45)
- Statistics (3)
- Computer Science (3)
- Genomics (7)
- Data science (8)
- Open science (4)
- Project management (1)
- Neuroethics (3)