Lesson type
Difficulty level
This lesson describes the fundamentals of genomics, from central dogma to design and implementation of GWAS, to the computation, analysis, and interpretation of polygenic risk scores.
Difficulty level: Intermediate
Duration: 1:28:16
Speaker: : Dan Felsky
This is a hands-on tutorial on PLINK, the open source whole genome association analysis toolset. The aims of this tutorial are to teach users how to perform basic quality control on genetic datasets, as well as to identify and understand GWAS summary statistics.
Difficulty level: Intermediate
Duration: 1:27:18
Speaker: : Dan Felsky
This is a tutorial on using the open-source software PRSice to calculate a set of polygenic risk scores (PRS) for a study sample. Users will also learn how to read PRS into R, visualize distributions, and perform basic association analyses.
Difficulty level: Intermediate
Duration: 1:53:34
Speaker: : Dan Felsky
This lesson is an overview of transcriptomics, from fundamental concepts of the central dogma and RNA sequencing at the single-cell level, to how genetic expression underlies diversity in cell phenotypes.
Difficulty level: Intermediate
Duration: 1:29:08
Speaker: : Shreejoy Tripathy
This is a tutorial introducing participants to the basics of RNA-sequencing data and how to analyze its features using Seurat.
Difficulty level: Intermediate
Duration: 1:19:17
Speaker: : Sonny Chen
This tutorial demonstrates how to perform cell-type deconvolution in order to estimate how proportions of cell-types in the brain change in response to various conditions. While these techniques may be useful in addressing a wide range of scientific questions, this tutorial will focus on the cellular changes associated with major depression (MDD).
Difficulty level: Intermediate
Duration: 1:15:14
Speaker: : Keon Arbabi
Similarity Network Fusion (SNF) is a computational method for data integration across various kinds of measurements, aimed at taking advantage of the common as well as complementary information in different data types. This workshop walks participants through running SNF on EEG and genomic data using RStudio.
Difficulty level: Intermediate
Duration: 1:21:38
Speaker: : Dan Felsky
This lesson continues from part one of the lecture Ontologies, Databases, and Standards, diving deeper into a description of ontologies and knowledg graphs.
Difficulty level: Intermediate
Duration: 50:18
Speaker: : Jeff Grethe
This lecture describes how to build research workflows, including a demonstrate using DataJoint Elements to build data pipelines.
Difficulty level: Intermediate
Duration: 47:00
Speaker: : Dimitri Yatsenko
This lesson characterizes different types of learning in a neuroscientific and cellular context, and various models employed by researchers to investigate the mechanisms involved.
Difficulty level: Intermediate
Duration: 3:54
Speaker: : Dan Goodman
In this lesson, you will learn about different approaches to modeling learning in neural networks, particularly focusing on system parameters such as firing rates and synaptic weights impact a network.
Difficulty level: Intermediate
Duration: 9:40
Speaker: : Dan Goodman
This tutorial provides instruction on how to simulate brain tumors with TVB (reproducing publication: Marinazzo et al. 2020 Neuroimage). This tutorial comprises a didactic video, jupyter notebooks, and full data set for the construction of virtual brains from patients and health controls.
Difficulty level: Intermediate
Duration: 10:01
Course:
This lesson provides an overview of Jupyter notebooks, Jupyter lab, and Binder, as well as their applications within the field of neuroimaging, particularly when it comes to the writing phase of your research.
Difficulty level: Intermediate
Duration: 50:28
Speaker: : Elizabeth DuPre
Course:
This lecture introduces you to the basics of the Amazon Web Services public cloud. It covers the fundamentals of cloud computing and goes through both the motivations and processes involved in moving your research computing to the cloud.
Difficulty level: Intermediate
Duration: 3:09:12
Speaker: : Amanda Tan & Ariel Rokem
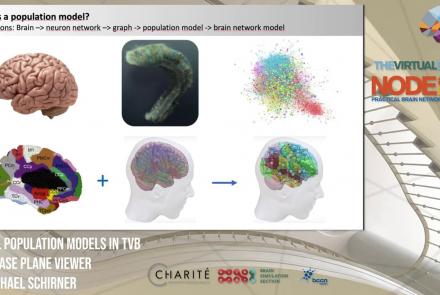
This lesson introduces population models and the phase plane, and is part of the The Virtual Brain (TVB) Node 10 Series, a 4-day workshop dedicated to learning about the full brain simulation platform TVB, as well as brain imaging, brain simulation, personalised brain models, and TVB use cases.
Difficulty level: Intermediate
Duration: 1:10:41
Speaker: : Michael Schirner
This lesson introduces TVB-multi-scale extensions and other TVB tools which facilitate modeling and analyses of multi-scale data.
Difficulty level: Intermediate
Duration: 36:10
Speaker: : Dionysios Perdikis
This lecture delves into cortical (i.e., surface-based) brain simulations, as well as subcortical (i.e., deep brain) stimulations, covering the definitions, motivations, and implementations of both.
Difficulty level: Intermediate
Duration: 39:05
Speaker: : Jil Meier
This lecture provides an introduction to entropy in general, and multi-scale entropy (MSE) in particular, highlighting the potential clinical applications of the latter.
Difficulty level: Intermediate
Duration: 39:05
Speaker: : Jil Meier
This lecture gives an overview of how to prepare and preprocess neuroimaging (EEG/MEG) data for use in TVB.
Difficulty level: Intermediate
Duration: 1:40:52
Speaker: : Paul Triebkorn
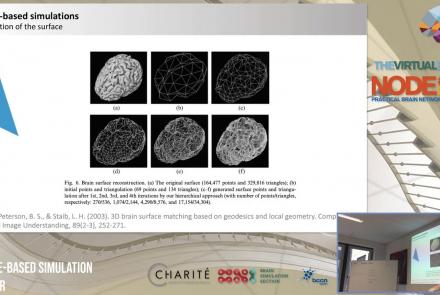
In this lecture, you will learn about various neuroinformatic resources which allow for 3D reconstruction of brain models.
Difficulty level: Intermediate
Duration: 1:36:57
Speaker: : Michael Schirner
Topics
- Artificial Intelligence (1)
- Provenance (1)
- EBRAINS RI (6)
- Animal models (1)
- Brain-hardware interfaces (1)
- Clinical neuroscience (20)
- General neuroscience (13)
- General neuroinformatics (1)
- Computational neuroscience (42)
- Statistics (5)
- Computer Science (2)
- (-) Genomics (7)
- Data science (8)
- (-) Open science (3)
- Project management (1)
- Neuroethics (3)