Lesson type
Difficulty level
Course:
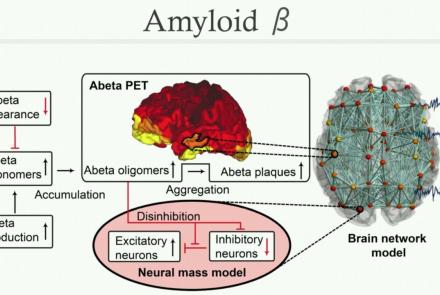
In this lecture we will focus on a paper called The Virtual Epileptic Patient: Individualized whole-brain models of epilepsy spread. We will have a closer look at the equations of the epileptor model and particular the epileptogenicity index, which controls the excitability of each brain region. Subsequently, we will begin to setup the epileptogenic zone in our own brain network model with TVB.
Difficulty level: Beginner
Duration: 6:25
Speaker: : Paul Triebkorn
Course:
After introducing the local epileptor model in the previous two videos, we will now use it in a large-scale brain simulation. We again focus on the paper The Virtual Epileptic Patient: Individualized whole-brain models of epilepsy spread. Two simulations with different epileptogenicity across the network are visualized to show the difference in seizure spread across the cortex.
Difficulty level: Beginner
Duration: 6:36
Speaker: : Paul Triebkorn
Course:
This lecture gives an overview on the article Individual brain structure and modelling predict seizure propagation, in which 15 subjects with epilepsy were modelled to predict individual epileptogenic zones. With the TVB GUI we will model seizure spread and the effect of lesioning the connectome. The impact of cutting edges in the network on seizure spreading will be visualized.
Difficulty level: Beginner
Duration: 9:39
Speaker: : Paul Triebkorn
This lecture presents the Graphical (GUI) and Command Line (CLI) User Interface of TVB. Alongside with the speakers, explore and interact with all means necessary to generate, manipulate and visualize connectivity and network dynamics.
Difficulty level: Beginner
Duration: 1:02:16
Speaker: : Paula Popa & Mihai Andrei
This lecture presents two recent clinical case studies using TVB: stroke recovery and dementia (due to Alzheimer’s Disease (AD)). Using a multi-scale neurophysiological model based on empirical multi-modal neuroimaging data, we show how local and global biophysical parameters characterize changes in individualized patient-specific brain dynamics, predict recovery of motor function for stroke patients, and correlate with individual differences in cognition for AD patients.
Difficulty level: Intermediate
Duration: 32:11
Speaker: : Randy McIntosh
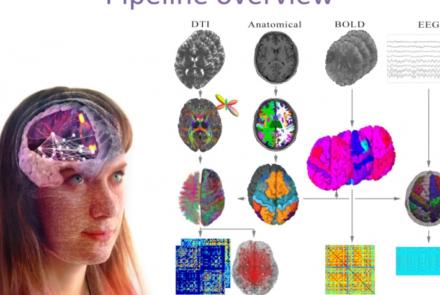
This tutorial demonstrates how to use the image processing pipeline with the HBP collab.
Difficulty level: Beginner
Duration: 5:55
Speaker: : M. Schirner, P. Triebkorn, P. Ritter
This tutorial provides instruction on how to use the TVB-NEST toolbox co-simulation in HBP collab.
Difficulty level: Beginner
Duration: 3:11
In this tutorial, you will learn how to use TVB-NEST toolbox on your local computer.
Difficulty level: Beginner
Duration: 2:16
This tutorial provides instruction on how to perform multi-scale simulation of Alzheimer's disease on The Virtual Brain Simulation Platform.
Difficulty level: Beginner
Duration: 29:08
This presentation accompanies the paper entitled: An automated pipeline for constructing personalized virtual brains from multimodal neuroimaging data (see link below to download publication).
Difficulty level: Beginner
Duration: 4:56
This lesson consists of a supplementary video for the publication: Inferring multi-scale neural mechanisms with brain network modelling.
Difficulty level: Beginner
Duration: 3:06
Course:
The Mouse Phenome Database (MPD) provides access to primary experimental trait data, genotypic variation, protocols and analysis tools for mouse genetic studies. Data are contributed by investigators worldwide and represent a broad scope of phenotyping endpoints and disease-related traits in naïve mice and those exposed to drugs, environmental agents or other treatments. MPD ensures rigorous curation of phenotype data and supporting documentation using relevant ontologies and controlled vocabularies. As a repository of curated and integrated data, MPD provides a means to access/re-use baseline data, as well as allows users to identify sensitized backgrounds for making new mouse models with genome editing technologies, analyze trait co-inheritance, benchmark assays in their own laboratories, and many other research applications. MPD’s primary source of funding is NIDA. For this reason, a majority of MPD data is neuro- and behavior-related.
Difficulty level: Beginner
Duration: 55:36
Speaker: : Elissa Chesler
Course:
This lesson provides a demonstration of GeneWeaver, a system for the integration and analysis of heterogeneous functional genomics data.
Difficulty level: Beginner
Duration: 25:53
Speaker: :
This tutorial provides instruction on how to simulate brain tumors with TVB (reproducing publication: Marinazzo et al. 2020 Neuroimage). This tutorial comprises a didactic video, jupyter notebooks, and full data set for the construction of virtual brains from patients and health controls.
Difficulty level: Intermediate
Duration: 10:01
The tutorial on modelling strokes in TVB includes a didactic video and jupyter notebooks (reproducing publication: Falcon et al. 2016 eNeuro).
Difficulty level: Intermediate
Duration: 7:43
This tutorial covers how to import appropriate data into The Virtual Brain, as well as how to begin constructing detailed brain models.
Difficulty level: Intermediate
Duration: 23:03
Speaker: : Patrik Bey
In this tutorial, you will learn how to run a typical TVB simulation.
Difficulty level: Intermediate
Duration: 1:29:13
Speaker: : Paul Triebkorn
This tutorial introduces The Virtual Mouse Brain (TVMB), walking users through the necessary steps for performing simulation operations on animal brain data.
Difficulty level: Intermediate
Duration: 42:43
Speaker: : Patrik Bey
In this tutorial, you will learn the necessary steps in modeling the brain of one of the most commonly studied animals among non-human primates, the macaque.
Difficulty level: Intermediate
Duration: 1:00:08
Speaker: : Julie Courtiol
This lecture provides an introduction to entropy in general, and multi-scale entropy (MSE) in particular, highlighting the potential clinical applications of the latter.
Difficulty level: Intermediate
Duration: 39:05
Speaker: : Jil Meier
Topics
- Standards and Best Practices (2)
- Notebooks (2)
- Clinical neuroinformatics (2)
- Provenance (2)
- Artificial Intelligence (1)
- Digital brain atlasing (3)
- Neuroimaging (36)
- Optogenetics (1)
- Standards and best practices (1)
- Tools (20)
- Workflows (3)
- protein-protein interactions (1)
- Extracellular signaling (1)
- Animal models (1)
- Assembly 2021 (1)
- Brain-hardware interfaces (12)
- (-) Clinical neuroscience (1)
- Repositories and science gateways (1)
- Resources (1)
- General neuroscience (11)
- Phenome (1)
- (-) Computational neuroscience (80)
- Statistics (5)
- Computer Science (4)
- Genomics (28)
- Data science (13)
- Open science (18)
- Education (1)
- Neuroethics (1)